Abstract
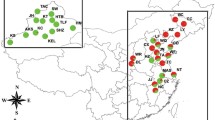
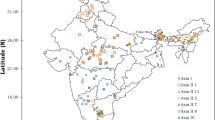
Whitefly Bemisia tabaci (Gennadius) is an important pest of cotton in India. The study of taxonomic diversity and its distribution on cotton is lacking. Such studies are necessary to identify the genetic groups of B. tabaci and its distribution on cotton in India. The proper identification of genetic groups and their distribution, which, ultimately, can lead to the timely development and utilization of management practices. The current study was undertaken to explore the genetic diversity and phylogeographic structure of B. tabaci from all the three major cotton growing zones representing different agro-climatic conditions of India. 290 partial mitochondrial Cytochrome Oxidase I (mt COI) sequences of whitefly population of six major cotton-growing states covering 22 districts were used in the analysis. Phylogenetic analysis revealed the presence of two monophyletic clades, Asia I and Asia II1 genetic groups represents South-Central India and North India respectively. Asia II1 is found more predominately distributed in the cotton leaf curl virus (CLCuV) disease-prone north Indian cotton-growing states. Higher genetic divergence (16.8–19.2%) was observed between the populations of Asia I and Asia II1 genetic group. Genetic differentiation analysis confirmed the phylogeographic structure of B. tabaci as isolated by distance. Our results in mapping the distribution of genetic groups in cotton ecosystems paved the way for the further studies and formulation of area-wide management practices for cotton whitely in India.




Similar content being viewed by others
Data availability
The authors declare that all the data used in the present study are original and generated from the study.
References
Ahmed MZ, De Barro PJ, Greeff JM, Ren S-X, Naveed M et al (2011) Genetic identity of the Bemisia tabaci species complex and association with high cotton leaf curl disease (CLCuD) incidence in Pakistan. Pest Manag Sci 67:307–317
AICCIP (2016–17) all India coordinated cotton improvement project report http://www.aiccip.cicr.org.in/CD_16_17/3_PC_Report.pdf
Ashfaq M, Hebert PDN, Mirza MS, Khan AM, Mansoor S, Shah GS, Zafar Y (2014) DNA barcoding of Bemisia tabaci Complex (Hemiptera: Aleyrodidae) reveals southerly expansion of the dominant whitefly species on cotton in Pakistan. PLoS One 9(8):e104485. https://doi.org/10.1371/journal.pone.0104485
Banks GK, Colvin J, Chowda RV, Maruthi MN, Muniyappa V, Venkatesh HM, Kiran Kumar M, Padma AS, Beitia FJ, Seal SE (2001) First report of the Bemisia tabaci B biotype in India and an associated tomato leaf curl virus disease epidemic. Plant Dis 85:231
Berry SD, Fondong VN, Rey C, Rogan D, Fauquet CM, Brown JK (2004) Molecular evidence for five distinct Bemisia tabaci (Homoptera: Aleyrodidae) geographic haplotypes associated with cassava plants in sub-Saharan Africa. Ann Entomol Soc Am 97:852–859
Briddon WR (2003) Cotton leaf curl disease, a multi-component begomovirus complex. Mol Plant Pathol 4:427–434
Bridle JR, Vines TH (2007) Limits to evolution at range margins: when and why does adaptation fail? Trends Ecol Evol 22:140–147
Brown JK (2000) Molecular markers for the identification and global tracking of whitefly vector-begomovirus complexes. Virus Res 71:233–260
Byrne DN, Bellows TS Jr (1991) Whitefly biology. Annu Rev Entomol 36:431–457
Cahill M, Denholm I, Ross G, Gorman K, Johnston D (1996) Relationship between bioassay data and the simulated field performance of insecticides against susceptible and resistant Bemisia tabaci. Bull Entomol Res 86:109–116
Chaubey R, Andrew RJ, Naveen NC, Rajagopal R, Ahmad B, Ramamurthy VV (2015) Morphometric analysis of three putative species of Bemisia tabaci (Hemiptera: Aleyrodidae) species complex from India. Ann Entomol Soc Am 108(4):600–612
Chowda-Reddy RV, Kirankumar M, Seal SE, Muniyappa V, Valand GB, Govindappa MR, Colvin J (2012) Bemisia tabaci phylogenetic groups in India and the relative transmission efficacy of tomato leaf curl Bangalore virus by an indigenous and an exotic population. J Integr Agric 11:235–248
Clement M, Snell Q, Walker P, Posada D, Crandall K (2002) TCS: Estimating gene genealogies. Parallel and Distributed Processing Symposium. Int Proc 2:84
Díaz F, Endersby NM, Hoffmann AA (2015) Genetic structure of the whitefly Bemisia tabaci populations in Colombia following a recent invasion. Insect Sci 22(4):483–494
Dinsdale A, Cook L, Riginos C, Buckley YM, De Barro PJ (2010) Refined global analysis of Bemisia tabaci (Gennadius) (Hemiptera: Sternorrhyncha: Aleyrodoidea) mitochondrial CO1 to identify species level genetic boundaries. Ann Entomol Soc Am 103:196–208
Ellango R, Singh ST, Rana VS, Priya G, Raina H, Chaubey R, Naveen NC, Mahmood R, Ramamurthy VV, Asokan R, Rajagopal R (2015) Distribution of Bemisia tabaci genetic groups in Indian. Environ Entomol 44(4):1258–1264
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol Ecol Resour 10:564–567
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinformatics Online 1:47–50
Frohlich DH, Torres-Jerez I, Bedford ID, Markham G, Brown JK (1999) A phylogeographical analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Mol Ecol 8:1683–1691
Gill RJ, Brown JK (2010) Systematics of Bemisia and Bemisia relatives: can molecular techniques solve the Bemisia tabaci complex conundrum-a taxonomist’s viewpoint. In: Stansly PA, Naranjo SE (eds) Bemisia: bionomics and management of a global pest. Springer, New York, pp 5–29
Gueguen G, Vavre F, Gnankine O, Peterschmitt M, Charif D, Chiel E, Gottlieb Y, Ghanim M, Fein EZ, Fleury F (2010) Endosymbiontmetacommunities, mtDNA diversity and the evolution of the Bemisiatabaci (Hemiptera: Aleyrodidae) species complex. Mol Ecol 19:4365–4376. https://doi.org/10.1111/j.1365-294X.2010.04775.x
Hampe A, Petit RJ (2005) Conserving biodiversity under climate change: the rear edge matters. Ecol Lett 8:461–467
Hsieh CH, Wang CH, Ko CC (2006) Analysis of Bemisia tabaci (Hemiptera: Aleyrodidae) species complex and distribution in eastern Asia based on mitochondrial DNA markers. Ann Entomol Soc Am 99:768–775
Hu J, De Barro P, Zhao H, Wang J, Nardi F, Liu SS (2011) An extensive field survey combined with a phylogenetic analysis reveals rapid and widespread invasion of two alien whiteflies in China. PLoS One 6:e16061
Jones RD (2003) Plant viruses transmitted by whiteflies. Eur J Plant Pathol 109:195–219
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Kellermann V, van Heerwaarden B, Sgrò CM, Hoffmann AA (2009) Fundamental evolutionary limits in ecological traits drive Drosophila species distributions. Science 325:1244–1246
Khatun MF, Hemayet Jahan SM, Lee S, Lee K-Y (2018) Genetic diversity and geographic distribution of the Bemisia tabaci species complex in Bangladesh. Acta Trop 187:28–36
Kranthi KR (2015) Whitefly –the black story. Cotton Stat News 23:1–3
Kranthi KR, Jadhav DR, Kranthi S, Wanjari RR, Ali SS, Russell DA (2002) Insecticide resistance in five major insect pests of cotton in India. Crop Prot 21:449–460
Kumar S, Stecher G, Tamura K (2016a) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Kumar NR, Chang JC, Narayanan MB, Ramasamy S (2016b) Phylogeographical structure in mitochondrial DNA of whitefly, Bemisia tabaci Gennadius (Hemiptera: Aleyrodidae) in southern India and Southeast Asia. Mitochondrial DNA Part A 28:621–631. https://doi.org/10.3109/24701394.2016.1160075
Legg J, French R, Rogan D, Okao-Okuja G, Brown JK (2002) A distinct Bemisia tabaci (Gennadius) (Hemiptera: Sternorrhyncha: Aleyrodidae) genotype cluster is associated with the epidemic of severe cassava mosaic virus disease in Uganda. Mol Ecol 11:1219–1229
Li Q, Deng J, Chen C, Zeng L, Lin X, Cheng Z, Qiao G, Huang X (2020) DNA barcoding subtropical aphids and implications for population differentiation. Insects 11:11
Liu S-s, Colvin J, De Barro PJ (2012) Species Concepts as Applied to the Whitefly Bemisia tabaci Systematics: How Many Species Are There? J Integr Agric 11(2):176–186
McKenzie CL, Bethke JA, Byrne FJ, Chamberlin JR, Dennehy TJ, Dickey AM, Gilrein D, Hall PM, Ludwig S, Oetting RD, Osborne LS, Schmale L, Shatters RG Jr (2012) Distribution of Bemisia tabaci (Hemiptera: Aleyrodidae) biotypes in North America after the Q invasion. J Econ Entomol 105:753–766
Misra CS, Lambda SK (1929) The cotton whitefly (Bemisia gossypiperda sp.). Bulletin of agricultural research, Inst. Pusa 196:1–7
Naveed M, Salam A, Saleem MA (2007) Contribution of cultivated crops, vegetable weeds and ornamental plants in harbouring of Bemisia tabaci (Homoptera: Aleyrodidae) and associated parasitoids (Hymenoptera: Aphelinidae). J Pest Sci 80:191–197
Naveen NC, Chaubey R, Kumar D, Rebijith KB, Rajagopal R, Subrahmanyam B, Subramaniana S (2017) Insecticide resistance status in the whitefly, Bemisia tabaci genetic groups Asia-I, Asia-II-1 and Asia-II-7 on the Indian subcontinent. Sci Rep 7:40634
Prabhulinga T, Rameash K, Madhu TN, Vivek S, Suke R (2017) Maximum entropy modelling for predicting the potential distribution of cotton whitefly Bemisia tabaci (Gennadius) in North India. J Entomol Zool Stud 5(4):1002–1006
Prabhulinga T, Gawande SP, Kranthi S, Monga D, Kumar R, Kranthi KR (2019) Status of cotton leaf curl virus disease and its vector whitefly in North India: a survey report. J Entomol Zool Stud 7(3):183–186
Prasanna HC, Kanakala S, Archana K, Jyothsna P, Varma RK, Malathi VG (2015) Cryptic species composition and genetic diversity within Bemisia tabaci complex in soybean in India revealed by mtCOI DNA sequence. J Integr Agric 14(9):1786–1795
Puillandre N, Lambert A, Brouillet S, Achaz G (2012) ABGD, automatic barcode gap discovery for primary species delimitation. Mol Ecol 21(8):1864–1877
Rana VS, Singh ST, Priya NG, Kumar J, Rajagopal R (2012) Arsenophonus GroEL interacts with CLCuV and is localized in the midgut and salivary gland of whitefly B. tabaci. PLoS One 7:e42168
Roopa HK, Asokan R, Rebijith KB, Ranjitha H, Hande RM, Krishna Kumar NK (2015) Prevalence of a new genetic group, MEAM-K, of the whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) in Karnataka, India, as evident from mt COI sequences. Fla Entomol 98(4):1062–1071
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol Biol Evol 34(12):3299–3302. https://doi.org/10.1093/molbev/msx248
Sethi A, Dilawari VK (2008) Spectrum of insecticide resistance in whitefly from upland cotton in the Indian subcontinent. J Entomol 5:138–147
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87:651–701
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3):585–595
Tyagi K, Kumar V, Singha D, Chandra K, Laskar BA, Kundu S, Chakraborty R, Chatterjee S (2017) DNA barcoding studies on Thrips in India: cryptic species and species complexes. Sci Rep 7:4898
Wright S (1978) Evolution and the genetics of populations, Vol. 4. Variability Within and Among Natural Populations. University of Chicago Press, Chicago
Xu J, Lin KK, Liu SS (2011) Performance on different host plants of an alien and an indigenous Bemisia tabaci from China. J Appl Entomol 135:771–779
Bedford ID, Briddon RW, Brown JK, Rosell RC, Markham PG (1994) Geminivirus transmission and biological characterisation of Bemisia tabaci (Genn.) biotypes from different geographic regions. Ann Appl Biol 125:311–325
Funding
Indian Council of Agricultural Research (ICAR)-Central Institute for Cotton Research (CICR), Nagpur, and National Initiative on Climate Resilient Agriculture (NICRA), India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable, as the research didn’t involve human/animals.
Consent to participate
The authors are equally contributed to the present study.
Consent for publication
All the authors of the manuscript declare their consent for publication.
Code availability
MEGA 7, PopART, DnaSP v6.10.01 and Arlequin v3.5 software were used for the analysis.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
T, P., Kranthi, S., P, R.K. et al. Mitochondrial COI based genetic diversity and phylogeographic structure of whitefly Bemisia tabaci (Gennadius) on cotton in India. Int J Trop Insect Sci 41, 1543–1554 (2021). https://doi.org/10.1007/s42690-020-00354-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42690-020-00354-x