Abstract
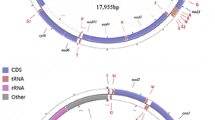
The pistachio psyllid, Agonoscena pistaciae, is one of the most deleterious pests of pistachio trees. Deciphering genetic diversity and population structure is a prerequisite for developing efficacious suppression protocols and successful management strategies for any insect pest including A. pistaciae. Mitochondrial genome sequencing furnishes valuable information about insect evolution and diversity within and among populations. Here, we assembled the first complete mitochondrial genome of the pistachio psyllid high-throughput next generation sequencing. The circular mitochondrial genome of A. pistaciae was 15,346 bp in length encoding 37 genes, including 13 protein-coding genes (PCGs), 2 rRNA genes and 22 tRNA genes, with a long non-coding region (putative control regions). The A. pistaciae mitogenome was in same typical set and arrangement as that of the ancestral insect. The mitochondrial genome of A. pistaciae was compared to all 6 publically available mitogenomes of other Psylloidea species. The conservation of the gene arrangements and identical gene ordering among all psyllid species could suggest a structurally common ancestral mitogenome shared across Psylloidea. Phylogenetic analysis based on the 13 PCGs conformed the existing morphological based taxonomy and showed the clustering of the two Aphalaridae. Whole mitochondrial genome of A. pistaciae could be utilized in subsequent population genetic studies of the species and also add to a deeper understanding of the phylogenetic research links within the subfamily Psylloidea.





Similar content being viewed by others
Data availability
All sequence data are available through GenBank Agonoscena Mitochondrial genome (PRJNA637689), the annotated mitochondrial genome (MT576697) and the illumine short reads (SRA) (SRR 11942466 & SRR 11942467) accession numbers.
References
Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF (2013) MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol 69:313–319
Bouckaert R, Heled J, Kuhnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10:e1003537
Burckhardt D, Ouvrard DA (2012) Revised classification of the jumping plant-lice (Hemiptera: Psylloidea). Zootaxa 3509:1–34
Camerson SL (2014) Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol 59:95–117
Cantacessi C, Jex AR, Hall RS, Young ND, Campbell BE, Joachim A, Nolan MJ, Abubucker S, Sternberg PW, Ranganathan S, Mitreva M, Gasser RBA (2010) Practical, bioinformatic workflow system for large data sets generated by next-generation sequencing. Nucleic Acids Res 38(17):e171
Chen J, Liang G (2016) The complete mitochondrial genome sequence of Bactericera cockerelli and comparison with three other Psylloidea species. PLoS One 11(5):e0155318. https://doi.org/10.1371/journal.pone.0155318
Chen S, Zhou Y, Chen Y, Gu J (2018) Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17):i884–i890
Cho G, Burckhardt D, Lee S (2017) On the taxonomy of Korean jumping plant-lice (Hemiptera: Psylloidea). Zootaxa 4238(4):531–561
Darling AE, Mau B, Perna NT (2010) Progressive mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. https://doi.org/10.1371/journal.pone.0011147
Heled J, Drummond AJ (2010) Bayesian inference of species trees from multilocus data. Mol Biol Evol 27:570–580
Herd KE, Barker SC, Shao R (2015) The mitochondrial genome of the chimpanzee louse, Pediculus schaeffi: insights into the process of mitochondrial genome fragmentation in the blood-sucking lice of great apes. BMC Genomics 16(1):661
Jeyaprakash A, Hoy MA (2009) First divergence time estimate of spiders, scorpions, mites and ticks (subphylum: Chelicerata) inferred from mitochondrial phylogeny. Exp Appl Acarol 47(1):1–18
Jiang F, Pan X, Li X, Yu Y, Zhang J, Jiang H, Dou L, Zhu S (2016) The first complete mitochondrial genome of Dacus longicornis (Diptera: Tephritidae) using next-generation sequencing and mitochondrial genome phylogeny of Dacini tribe. Sci Rep 6:36426. https://doi.org/10.1038/srep36426
Karimi S, Hosseini R, Farahpour A, Aalami A (2014) Application of RAPD in comparison of diversity of commonpistachio psylla (Agonoscena pistaciae Burckhardt & Lauterer) populations in some northern and southern regions of Kerman province. Plant Pest Res 4(1):21–34 (In Persian)
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Li H, Liu H, Shi A, Stys P, Zhou X, Cai W (2012) The complete mitochondrial genome and novel gene arrangement of the unique-headed bug Stenopirates sp. (Hemiptera: Enicocephalidae). PLoS One 7(1):e29419
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Martoni F, Armstrong KF (2019) Acizzia errabunda sp. nov. and Ctenarytaina insularis sp.nov.: descriptions of two new species of psyllids (Hemiptera:Psylloidea) discovered on exotic host plants in New Zealand. PLoS ONE 14(4):e0214220
Mehrnejad MR (2002) Bionomics of the common pistachio psylla, Agonoscena pistaciae, in Iran. Acta Hortic 591:535–539
Mehrnejad MR (2003) Pistachio Psylla and other major Psyllids of Iran. Ministry of Jihad-e-Agriculture, Tehran
Mehrnejad MR (2010) Potential biological control agents of the common pistachio psylla, Agonoscena pistaciae, a review. Entomofauna 31(21):317–340
Mehrnejad MR (2014) The psyllids of pistachio trees in Iran. Acta Hortic 1028:191–194
Mostafavi M, Lashkari M, Iranmanesh S, Mansouri SM (2017) Variation in populations of common pistachio psyllid, Agonoscena pistaciae (Hem.: Psyllidae), with different chemical control levels: narrower wing shape in the stressed environment. J Crop Prot 6(3):353–362
Nezam Kheirabadi M, Dhami MK, Fekrat L, Lashkari MR, Pramual P (2020) Genetic structure and diversity of the common pistachio psylla, Agonoscena pistaciae Burckhardt & Lauterer, (Hemiptera: Aphalaridae) in Iran. Manusript submitted for publication
Ouvrard D. (2020) Psyl’list— The World Psylloidea Database. http://www.hemiptera-databases.com/psyllist- searched on the 5th of May 2020
Perilla-Henao LM, Casteel CL (2016) Vector-borne bacterial plant pathogens: interactions with hemipteran insects and plants. Front Plant Sci 7:1163. https://doi.org/10.3389/fpls.2016.01163
Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41:353–358
Que S, Yu L, Xin T, Zou Z, Hu L, Xia B (2016) Complete mitochondrial genome of Cacopsylla coccinae (Hemiptera: Psyllidae). Mitochondrial DNA A DNA Mapp Seq Anal 27(5):3169–3170
Rambaut A. (2012) FigTree (version 1.4.3), http://tree.bio.ed.ac.uk/software/figtree/
Song F, Li H, Jiang P, Zhou X, Liu J, Sun C, Volger AP, Cai W (2016) Capturing the phylogeny of Holometabola with mitochondrial genome data and Bayesian site-heterogeneous mixture models. Genome Biol Evol 8(5):1411–1426
Thao ML, Baumann L, Baumann P (2004) Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha). BMC Evol Biol 4(1):25.13
Wang HL, Xiao N, Yang J, Wang XW, Colvin J, Liu SS (2016) The complete mitochondrial genome of Bemisia afer (Hemiptera: Aleyrodidae). Mitochondrial DNA 27(2):98–99
Wang N, Pierson EA, Setubal JC, Xu J, Levy JG, Zhang Y, Li J, Rangel LT, Martins J Jr (2017) The Candidatus Liberibacter-host Interface: insights into pathogenesis mechanisms and disease control. Annu Rev Phytopathol 55:451–482. https://doi.org/10.1146/annurev-phyto-080516-035513
Wang AY, Peng YQ, Harder LD, Huang JF, Yang DR, Zhang DY, Liao WJ (2019) The nature of interspecific interactions and co-diversification patterns, as illustrated by the fig microcosm. New Phytol 224:1304–1315
Williams ST, Foster PG, Littlewood DTJ (2014) The completemitochondrial genome of a turbinid vetigastropod from MiSeq Illu-mina sequencing of genomic DNA and steps towards a resolved gas-tropod phylogeny. Gene 533:38–47
Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. Int Rev Cytol 141:173–216
Wu F, Cen Y, Wallis CM, Trumble JT, Prager S, Yokomi R, Zheng Z, Deng X, Wang Y, Huang XL, Qiao GX (2014) The complete mitochondrial genome of Cervaphis quercus (Insecta: Hemiptera: Aphididae: Greenideinae). Insect sci 21(3):278–290
Wu F, Cen Y, Deng X, Zheng Z, Chen J, Liang G (2016a) The complete mitochondrial genome sequence of Diaphorina citri (Hemiptera: Psyllidae). Mitochondrial DNA B 1(1):239–240
Wu F, Cen Y, Wallis CM, Trumble JT, Prager S, Yokomi R, Zheng Z, Deng X, Chen J, Liang G (2016b) The complete mitochondrial genome sequence of Bactericera cockerelli and comparison with three other Psylloidea species. PLoS One 11(5):e0155318. https://doi.org/10.1371/journal.pone.0155318
Zhang QL, Guo ZL, Yuan ML (2016a) The complete mitochondrial genome of Poratrioza sinica (Insecta: Hemiptera: Psyllidae). Mitochondrial DNA Part A 27(1):734–735
Zhang S, Luo J, Wang C, Lv L, Li C, Jiang W, Cui J, Rajput LB (2016b) Complete mitochondrial genome of Aphis gossypii glover (Hemiptera: Aphididae). Mitochondrial DNA Part A 27(2):854–855
Zhang Y, Feng S, Fekrat L, Jiang F, Khathutshelo M, Li Z (2019) The first two complete mitochondrial genome of Dacus bivittatus and Dacus ciliatus (Diptera: Tephritidae) by next-generation sequencing and implications for the higher phylogeny of Tephritidae. Int J Biol Macromol 140:469–476
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 13 kb)
Rights and permissions
About this article
Cite this article
Fekrat, L., Zakiaghl, M., Dhami, M.K. et al. De novo assembly and comparative analysis of the complete mitochondrial genome sequence of the pistachio psyllid, Agonoscena pistaciae (Hemiptera: Aphalaridae). Int J Trop Insect Sci 41, 1387–1396 (2021). https://doi.org/10.1007/s42690-020-00332-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42690-020-00332-3