Abstract
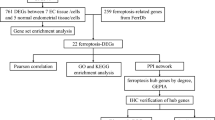
Breast cancer (BC), Endometrial cancer (EC) and Ovarian cancer are devastating diseases among women, because the ratio of death is very high for these three cancers. The arising point of endometrial cancer is uterus, which is a pelvic organ and development of fatal occur in uterus. Among all the risk factors of endometrial cancer, the prominent position is held by breast cancer, because a significant amount of molecular pathways, as well as seed genes, are linked with one another. There are two ovaries in the reproductive system of female and the positions of these ovaries are in both side of uterus. Ovary is the place, where ovarian cancer arises from. The present study attempts to find the common gene among BC, EC and OC. Reduction of gene rate is achieved through the preprocessing and filtering process. Protein–protein interaction (PPIs) network is designed for 335 common gene generated from gene mining process. Topological analysis finally provides ten common genes necessary for analysis of pathways, Gene regulatory Network (GRN), co-expression, physical interaction network. Gene ontology analysis generates better understanding of biological process, cellular component and molecular functioning. Interaction of proteins with drug molecules comes up with efficient drug design for this research.














Similar content being viewed by others
Abbreviations
- BC:
-
Breast cancer
- EC:
-
Endometrial cancer
- OC:
-
Ovarian cancer
- NCBI:
-
National Center of Biotechnology Information
- GO:
-
Gene ontology
- CTD:
-
Comparative toxicogenomics database
- GRN:
-
Gene regulatory network
References
Alexeyenko A, Lee W, Pernemalm M, Guegan J, Dessen P, Lazar V, Lehtiö J, Pawitan Y (2012) Network enrichment analysis: extension of gene-set enrichment analysis to gene networks. BMC Bioinform 13(1):226
Antoni MH, Lutgendorf SK, Cole SW, Dhabhar FS, Sephton SE, McDonald PG, Stefanek M, Sood AK (2006) The influence of bio-behavioural factors on tumour biology: pathways and mechanisms. Nat Rev Cancer 6(3):240–248
Arem H, Irwin ML (2013) Obesity and endometrial cancer survival: a systematic review. Int J Obes 37(5):634–639
Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M (2008) Computing topological parameters of biological networks. Bioinformatics 24(2):282–284
Barabási AL, Gulbahce N, Loscalzo J (2011) Network medicine: a network-based approach to human disease. Nat Rev Genet 12(1):56–68
Cha S, Imielinski MB, Rejtar T, Richardson EA, Thakur D, Sgroi DC, Karger BL (2010) In situ proteomic analysis of human breast cancer epithelial cells using laser capture microdissection: annotation by protein set enrichment analysis and gene ontology. Mol Cell Proteom 9(11):2529–2544
Chen JY, Kuo SJ, Liaw YP, Avital I, Stojadinovic A, Man YG, Mannion C, Wang J, Chou MC, Tsai HD, Chen ST (2014) Endometrial cancer incidence in breast cancer patients correlating with age and duration of tamoxifen use: a population based study. J Cancer 5(2):151
DeSantis C, Siegel R, Bandi P, Jemal A (2011) Breast cancer statistics. CA Cancer J Clin 61(6):408–418
Ding Y, Tang J, Guo F (2016) Identification of protein–protein interactions via a novel matrix-based sequence representation model with amino acid contact information. Int J Mol Sci 17(10):1623
Doms A, Schroeder M (2005) GoPubMed: exploring PubMed with the gene ontology. Nucleic Acids Res 33(suppl_2):W783–W786
Dossus L, Allen N, Kaaks R, Bakken K, Lund E, Tjonneland A, Olsen A, Overvad K, Clavel-Chapelon F, Fournier A, Chabbert-Buffet N (2010) Reproductive risk factors and endometrial cancer: the European prospective investigation into cancer and nutrition. Int J Cancer 127(2):442–451
García-Campos MA, Espinal-Enríquez J, Hernández-Lemus E (2015) Pathway analysis: state of the art. Front Physiol 6:383
Herrero J, Díaz-Uriarte R, Dopazo J (2003) Gene expression data preprocessing. Bioinformatics 19(5):655–656
Huang SM, Zhao H, Lee JI, Reynolds K, Zhang L, Temple R, Lesko LJ (2010) Therapeutic protein–drug interactions and implications for drug development. Clin Pharmacol Ther 87(4):497–503
Jeong H, Mason SP, Barabási AL, Oltvai ZN (2001) Lethality and centrality in protein networks. Nature 411(6833):41–42
Khatri P, Sirota M, Butte AJ (2012) Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol 8(2):e1002375
Kuhn M, Szklarczyk D, Franceschini A, Von Mering C, Jensen LJ, Bork P (2012) STITCH 3: zooming in on protein–chemical interactions. Nucleic Acids Res 40(D1):D876–D880
Leung HC, Xiang Q, Yiu SM, Chin FY (2009) Predicting protein complexes from PPI data: a core-attachment approach. J Comput Biol 16(2):133–144
Lussier Y, Borlawsky T, Rappaport D, Liu Y, Friedman C (2006) PhenoGO: assigning phenotypic context to gene ontology annotations with natural language processing. Biocomputing 2006:64–75
Montojo J, Zuberi K, Rodriguez H, Bader GD, Morris Q (2014) GeneMANIA: fast gene network construction and function prediction for Cytoscape. F1000Res 3:153
Oti M, Snel B, Huynen MA, Brunner HG (2006) Predicting disease genes using protein–protein interactions. J Med Genet 43(8):691–698
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics. CA 69(1):7–34
Song XY, Chen ZH, Sun XY, You ZH, Li LP, Zhao Y (2018) An ensemble classifier with random projection for predicting protein–protein interactions using sequence and evolutionary information. Appl Sci 8(1):89
Sumanasekera W, Beckmann T, Fuller L, Castle M, Huff M (2018) Epidemiology of ovarian cancer : risk factors and prevention. Biomed J Sci Tech Res 5:1–13
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613
Tang J, Rangayyan RM, Xu J, El Naqa I, Yang Y (2009) Computer-aided detection and diagnosis of breast cancer with mammography: recent advances. IEEE Trans Inf Technol Biomed 13(2):236–251
Tian C, Zhang X, He J, Yu H, Wang Y, Shi B, Han Y, Wang G, Feng X, Zhang C, Wang J (2014) An organ boundary‐enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol Syst Biol 10(10):755
Tieri P, Farina L, Petti M, Astolfi L, Paci P, Castiglione F (2019) Network inference and reconstruction in bioinformatics. pp 805–813
Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A, Siegel RL (2018) Ovarian cancer statistics (2018). CA Cancer J Clin 68(4):284–296
Urick ME, Bell DW (2019) Clinical actionability of molecular targets in endometrial cancer. Nat Rev Cancer 19(9):510–521
Walhout AJ (2011) Gene-centered regulatory network mapping. Methods Cell Biol 106:271–288 Academic Press
Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38(suppl_2):W214–W220
Weirauch MT (2011) Gene coexpression networks for the analysis of DNA microarray data. Appl Stat Netw Biol Methods Syst Biol 1:215–250
Yeung N, Cline MS, Kuchinsky A, Smoot ME, Bader GD (2008) Exploring biological networks with Cytoscape software. Curr Protoc Bioinform 23(1):8–13
Young TE (2012) Therapeutic drug monitoring–the appropriate use of drug level measurement in the care of the neonate. Clin Perinatol 39(1):25–31
Yu D, Lim J, Wang X, Liang F, Xiao G (2017) Enhanced construction of gene regulatory networks using hub gene information. BMC Bioinform 18(1):186
Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4(1):1–43
Zhang J, Huan J (2010) Comparison of chemical descriptors for protein–chemical interaction prediction. Int J Comput Biosci 1(1):13–21
Zhou G, Soufan O, Ewald J, Hancock RE, Basu N, Xia J (2019) NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucl Acids Res 47(W1):W234–W241
Acknowledgements
This manuscript has not been published yet and not even under consideration for publication elsewhere. The authors are grateful who have participated in this research work.
Funding
There is no funding for this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors have read the manuscript and approved this for submission as well as no competing interests.
Rights and permissions
About this article
Cite this article
Taz, T.A., Kawsar, M., Paul, B.K. et al. Computational analysis of regulatory genes network pathways among devastating cancer diseases. J Proteins Proteom 11, 63–76 (2020). https://doi.org/10.1007/s42485-020-00032-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42485-020-00032-z