Abstract
Cervical cancer is one type of gynaecological cancers and the majority of these complications of cervical cancer are associated to human papillomavirus infection. There are numerous risk factors associated with cervical cancer. It is important to recognize the significance of test variables of cervical cancer for categorizing the patients based on the results. This work intended to attain deeper understanding by applying machine learning techniques in R to analyze the risk factors of cervical cancer. Various types of feature selection techniques are explored in this work to determine about important attributes for cervical cancer prediction. Significant features are identified over various iterations of model training through several feature selection methods and an optimized feature selection model has been formed. In addition, this work aimed to build few classifier models using C5.0, random forest, rpart, KNN and SVM algorithms. Maximum possibilities were explored for training and performance evaluation of all the models. The performance and prediction exactness of these algorithms are conferred in this paper based on the outcomes attained. Overall, C5.0 and random forest classifiers have performed reasonably well with comprehensive accuracy for identifying women exhibiting clinical sign of cervical cancer.
Similar content being viewed by others
1 Introduction
Gynaecological cancers are those that develop in a woman’s reproductive tract and they are the most common type of cancers in women after breast cancer. Gynaecological cancers are very dangerous and lead to lessening the lifespan of women diagnosed with such type of cancers. Cervical cancer is one type of gynaecological cancer, other types are Ovarian cancer, Uterine cancer, Vaginal cancer and Vulvar cancer. There are different risk factors for each type of gynaecological cancers. Cervical cancer is the second most commonly identified cancer in women and representing 7.5% of all female cancer deaths all over the world [1]. Cervical cancer is malicious tumor that occurs when the cervix tissue cells begin to grow and reproduce abnormally without controlled cell division and cell death. If the tumor is malignant, its cell flow through the blood stream and spread to other parts of body, consequently those parts also get infected, and in maximum cases it can be prevented through early detection [2].
Generally medical dataset is provided with more attributes and missing values [3]. Identifying the relevant and important features for statistical model building is essential by way of optimization. It is apparent that Machine Learning (ML) methods are more beneficial in predictions, optimization related explorations and they have been extensively implemented in various types of cancer researches. The study [4] which discussed about various works relevant to cancer prediction/prognosis evidenced accurate results attainment by means of ML techniques. R is one of the most popular and widely-used software structures for statistics, data mining, and machine learning. The R packages offer an innovative, easy-to-use, and flexible domain-specific functions for machine learning experiments [5]. It supports classification, regression, clustering, and survival analysis with more modelling techniques. Accordingly, an efficient classifier model for cervical cancer prediction can be built by implementing ML methods in R and the correctness of the model can be estimated using various evaluation metrics to attain enhanced performance efficiency.
2 Related work
The studies on women cancer [6, 7] developed a prediction model by combining classifier methods and feature selection techniques in order to significantly improve predictive accuracy for breast cancer diagnosis and prognosis. The study on staging prediction in cervical cancer [8] aimed at identifying the most influential risk factors by using decision tree classifier and extracted the rules based on the signs and symptoms observed from the dataset. The study on cervical cancer data [9] applied RUS and ROS methods for balancing of data. Stability Selection (SS) method was used for feature selection. In this work, 190 instances including missing values (‘?’, Null) were removed from the raw dataset. So, there were 668 records in the raw dataset. Here, learning model was designed based on the combination of SS method and RF algorithm. The success of this model was tested on the RUS and ROS methods. The results showed that ROS based SS method more successful than RUS based SS method on this dataset and this work achieved 98% accuracy. Another work [10] on cervical cancer data used KNN algorithm and it has selected 25 features, decision tree classifier selected 17 features and random forest algorithm selected 11 features for prediction. This work concluded that KNN algorithm seems to be the best model with higher accuracy and AUC which is 0.822 as compared to Decision tree and random forest algorithms. But the number of training and test data samples selected from cervical cancer dataset were varied in each algorithm studied in this work. The study [11] to classify cervical cancer data applied over sampling, under sampling and combined sampling methods to handle the imbalanced data. This method selected six features as important and attained 97% accuracy with decision tree classifier. Observational studies have shown that the cervical cancer dataset considered in various works have removed the instances with missing values and less importance has been given in determining significant attributes. Subsequently, there is a challenge in dealing with missing values in dataset, determining precise attributes and accomplishing the results of higher prediction accuracy with optimization. Therefore, this work is intended to attain these challenges.
3 Feature selection
Feature selection is a process in which the features that contribute more to the estimated predictor variable are automatically selected from the data. Feature selection (FS) methods can be used in data pre-processing to accomplish effective data reduction and this is suitable for finding accurate data models [12,13,14]. Selecting appropriate features in the data are important, since irrelevant features can decrease the accuracy of many models [15]. We need not use every feature present in the data for creating an algorithm. We can train our algorithm with those features that are certainly important and it will authorize improved results than using complete set of features for the same algorithm.
3.1 Advantages of using feature selection
-
Allows the ML procedure to train the model more rapidly
-
Reduces model complexity with an ease of interpretation
-
Advances the precision of a model when the precise subset is selected
-
Decreases overfitting
3.2 Feature selection techniques
3 types, they are filter methods, wrapper methods and embedded methods.
3.2.1 Filter methods
Filter methods are normally used as a pre-processing stage. Here the features are selected based on their correlation with the outcome variable through statistical tests i.e. It measures the importance of features through their correlation with dependent variable. The feature selection process with filter methods is depicted in Fig. 1. Filter methods are considerably faster than wrapper methods.
3.2.2 Wrapper methods
In wrapper methods, subset of features is used for training a model. Based on the inferences gained from the preceding model, inclusion or removal of features from the subset can be decided. Thus, it measures the effectiveness of a subset of feature by means of training a model on it. Hence these methods are computationally higher. The wrapper method for feature selection process is represented in Fig. 2.
Some common examples of wrapper approaches are forward feature selection, backward feature elimination and recursive feature elimination.
Forward selection - It is an iterative method, initially there will not be any feature in the mode and in each iteration, new feature is added which best advances the model. This will be continued till an addition of a new variable does not advances the model performance.
Backward elimination - In this method, we begin with all the features and eliminates the minimum substantial feature at each iteration. This process is repeated until there is no progression is detected by eliminating the features.
Recursive feature elimination - It is an optimization algorithm and intends to attain the finest feature subset. It continually produces models and determine the finest or the worst feature at each repetition. It creates the subsequent model with the remaining features till entire features are explored. Then it organizes all the features with respect to their order of elimination.
3.2.3 Embedded methods
The attribute selection using embedded method is described in Fig. 3. This method combines the abilities of both the methods discussed earlier. It is executed by procedures which have their specific built-in feature selection methods.
Various study on cancer classification approach through wrapper-based feature selection [16] showed an excellent performance, not only at identifying relevant genes, but also with respect to the computational cost. Accordingly, wrapper methods are used in this work for feature selection to see whether the accuracy of the model can be improved through perceptively nominated subset of features rathe than using all features in the dataset.
4 Cervical cancer risk factors analysis and prediction
This work consists of four main stages which include the data preparation—cleaning of data, identification of significant test variables or predictors, model training/building of classifier model and performance evaluation.
4.1 Data preparation
The dataset used in this work is the openly accessible cervical cancer dataset from UCI Machine Learning Repository [17] which was gathered at Hospital Universitario de Caracas in Caracas, Venezuela. This dataset comprises medicinal histories of 858 patients with 36 attributes (32 input features and 4 target variables as Hinselmann, Schiller, Cytology, Biopsy). The attributes information of the dataset is given in Table 1.
It is essential to feed the right data to the machine learning processes for the problem to be solved, since these algorithms learn from data. After selecting the data, it should be pre-processed and transformed.
4.1.1 Data cleaning: dealing with missing values
This cervical cancer dataset is having lot of missing values. Sometimes missing values are a common existence, and we require an efficient approach for handling such information. A missing value can imply a number of different things in the data. The records with missing values can be ignored or they can be replaced with the variable mean for numerical attributes or by most frequent value in case of categorical attributes. When we applied the approach to remove the records with missing values the number of rows reduced to 737 from 858. Our aim is to reduce the number of features but not the number of records available in the dataset, hence the strategy of replacing the missing values with mean is used for numerical attributes.
The columns for STDs_cervical_condylomatosis, STDs_vaginal_condylomatosis, STDs_pelvic_inflam_disease, STDs_genital_herpes, STDs_molluscum_contagiosum, STDs_Hepatitis_B, STDs_HPV and STD_AIDS were removed, since there were 4 or less patient results for these features. Similarly, the features STDs_Time_since_first_diagnosis and STDs_Time_since_last_diagnosis which contained greater than 60% missing values (787 of 858) were also eliminated from the dataset. Subsequently, after removing all these columns the dataset comprised of 858 rows with 26 features.
4.1.2 Creation of a target feature
The values of four target variables Hinselmann, Schiller, Citology and Biopsy represent the results for cervical cancer exams. The histogram representation with four target variables is shown below in Fig. 4.
The data for these columns can be combined to create a single target feature called ‘Cancer’ as 27th feature. The advantage of combining all the four target variables is to confirm the possibility of the diagnosis. Higher values for this feature signify an increased likelihood of cervical cancer. If one diagnosis indicate that the patient has cancer, but the other three diagnosis attained different results then the possibility of the patient having cancer is doubtful. However, if all four diagnoses are showing that the patient does not have cancer, then the chances that the patient not having cancer is moderately high. The combined target variables representation is shown in Fig. 5.
So, it is determined that 87% of the patients do not have cancer. This can be our baseline.
4.2 Applying feature selection to data
When the dimensionality of the data increases the computational cost also increases exponentially. In the existence of several inappropriate features, learning models incline to overfit and convert as less efficient [18]. To overcome this problem, it is required to find a method to diminish the number of features in consideration. Feature subset selection works by eliminating the features that are redundant or not appropriate. During data cleaning process, ten columns were removed which had missing values and 27th feature ‘Cancer’ is added as target by combing other four target values. In feature selection process, this target variable has been used to find the important and relevant features. So, the dataset is now available with 858 rows, 27 attributes. (22 predictor variables +4 target variables + an additional combined target variable ‘Cancer’). In this work, various types of feature selection methods are explored using R tool to identify most significant and optimal features.
4.2.1 Recursive feature elimination (RFE)
It is evident from our understanding that the dataset is unbalanced, hence K-fold-cross-validation is required to attain better outcomes. RFE is a feature selection method that fits a model and eliminates the weakest features. Cross-validation is used with RFE to find the optimal number of features and finest ranking set of features are selected. In R tool, Recursive Feature Elimination—RFE procedure can be implemented using caret package. Initially the control function to be used in RFE algorithm should be defined. The random forest selection function over rfFuncs option in rfeControl function is stated here.
-
control < - rfeControl(functions = rfFuncs, method = ”cv”, number = 10)
Then RFE algorithm is implemented as follows.
-
rfe.train < - rfe(training_data[,1:22], training_data[,23], sizes = 1:10, rfeControl = control)
The original target variables in the dataset were removed and the procedure was implemented with predictor variables and newly added target variable. After the implementation of RFE algorithm the result has been plotted and variable importance chart has been obtained. The chart is depicted in Fig. 6.
rfe.train output has shown the following result.
The top 3 variables (out of 3):
Dx.HPV, | Dx.Cancer, | Dx |
Predictors(rfe.train) has revealed the following output.
predictors(rfe.train)
[1] | “Dx.HPV” | “Dx.Cancer” | “Dx” |
So RFE algorithm has predicted three features Dx.HPV, Dx.Cancer, Dx as important.
4.2.2 Boruta algorithm
All-relevant feature selection is a moderately new sub-field in the province of feature selection [19]. Boruta is an all relevant feature selection algorithm in R which functions as a wrapper algorithm around Random Forest. It makes a top-down search for appropriate features by associating original attributes’ importance with importance attainable at random, assessed using their permuted copies, and gradually rejecting inappropriate features. Boruta captures all features which are statistically significant to the target variable in some conditions.
Working Principle of Boruta Algorithm - The procedure of Boruta Algorithm is explained with sequence of phases.
-
(a)
Initially, it assigns randomness to the given dataset by making shuffled copies of all features (termed as shadow features).
-
(b)
Later, it trains a random forest classifier on the dataset and applies a feature ranking measure (Mean Decrease Accuracy) to estimate the relevance (higher mean value) of each feature.
-
(c)
On each iteration, it finds whether a real feature has a higher position than the best of its shadow features and continuously eliminates features which are estimated extremely insignificant.
-
(d)
The algorithm halts when all features gets confirmed or excluded or when it accomplishes a stated limit of random forest runs.
In Boruta, maxRuns is the number of times the algorithm is supposed to run. The higher the maxRuns the more selectively the variables can be picked. The default value is 100. Boruta check for all features which are either strongly or weakly pertinent to the target variable. With Boruta the dataset of missing values should not be used to identify significant variables. As the goal is to find the features (other than four target variables) which are all significant to decide the outcome as Cancer or Not, the same set of data which was used in RFE algorithm was used in Boruta also. After training our dataset with Boruta algorithm it produces the following output.
print(boruta.train)
-
Boruta performed 99 iterations in 58.59354 s.
-
5 attributes confirmed important: Dx, Dx.Cancer, Dx.HPV,
-
Smokes..packs.year., STDs.vulvo.perineal.condylomatosis;
-
7 attributes confirmed unimportant:
-
First.sexual.intercourse, Hormonal.Contraceptives, IUD,
-
IUD..years., Num.of.pregnancies and 2 more;
-
10 tentative attributes left: Age, Dx.CIN,
-
Hormonal.Contraceptives..years., Number.of.sexual.partners,
-
Smokes and 5 more;
Here the top three features were already selected by RFE algorithm. The variable importance chart of Boruta algorithm is portrayed in Fig. 7. The plot is shown for all the attributes taken into consideration. Blue boxplots represent minimal, average and maximum Z score of a shadow attribute. Red, yellow and green boxplots indicate the Z scores of rejected, tentative and confirmed attributes respectively.
In the process of deciding if a feature is important or not, some features may be marked by Boruta as ’Tentative’. These tentative attributes will be decided as confirmed or rejected by comparing the median Z score of the attributes with the median Z score of the best shadow attribute. After deciding on tentative attributes Boruta produced the following output for cervical cancer data and the chart is showed in Fig. 8.
getSelectedAttributes(final.boruta, withTentative = F)
-
[1]
“Smokes..packs.year.”
-
[2]
“Hormonal.Contraceptives..years.”
-
[3]
“STDs”
-
[4]
“STDs.vulvo.perineal.condylomatosis”
-
[5]
“STDs.syphilis”
-
[6]
“STDs..Number.of.diagnosis”
-
[7]
“Dx.Cancer”
-
[8]
“Dx.HPV”
-
[9]
“Dx”
Boruta algorithm has shown a much-improved result of variable importance as compared to the old feature selection method (RFE). In Boruta, it is easy to understand the results through the clear interpretation.
4.2.3 Simulated annealing (SA)
Simulated annealing is a search algorithm that allows a suboptimal solution to be accepted with an expectation that a better solution will be obtained at the end. This algorithm is used with an aim to get the optimal solution by producing a smaller number of feature subsets for evaluation [20]. It works by doing minor random changes to a preliminary solution and checks for the improvement in the performance. The optimal variables obtained for cervical cancer risk factors through Simulated Annealing (safs function) are shown below.
print(sa_obj$optVariables)
-
[1]
“Age” ”Smokes..years.”
-
[3]
“Hormonal.Contraceptives” ”STDs..number.”
-
[5]
“STDs..Number.of.diagnosis” ”Dx.Cancer”
-
[7]
“Dx.HPV”
This procedure has derived seven important features from cervical cancer dataset.
4.2.4 Feature selection with machine learning algorithms
There is an alternate way to accomplish feature selection is to consider variables most used by several Machine Learning (ML) algorithms the most to be significant. Initially ML algorithms learn the association between X’s and Y then based on the learning, different machine learning algorithms could probably end up using different variables to various degrees. Therefore, the variables that showed suitable in a tree-based algorithm like rpart, can turn out to be less valued in a regression-based model. Hence, all variables need not be equally appropriate to all algorithms. It is apparent that employing feature subset selection using wrapper approach in ML algorithms could enhance classification accuracy [6]. Hence this work is intended to apply feature subset selection with few ML algorithms to validate and compare their performance. Steps to find variable Importance from ML Algorithms are shown below.
-
The desired model should be trained through train() function using the caret package
-
Then varImp() function is applied for finding important features
Few of ML algorithms namely rpart, C5.0, svmRadial, knn, ctree and rf were applied in this work to decide about the features which are significant to attain reliable accuracy with optimization. All these ML algorithms are intended to train the model and the models built by these algorithms would be applied on test data. Hence, we decided to use the dataset with 26 predictor variables which included those four target variables (Schiller, Citology, Biopsy, Hinselmann) also. The models were trained with respect to the decisive target variable ‘Cancer’.
rpart()—The R implementation of CART algorithm is termed as RPART (Recursive Partitioning And Regression Trees). The rpart algorithm works by splitting the dataset recursively, until a predetermined termination condition is reached. At each step, the split is made based on the independent variable which allows major possible reduction in heterogeneity of the predicted variable. rpart method has shown the following output for the cervical cancer dataset considered in our work.
rpart variable importance (Output obtained in R)
only 20 most important variables shown (out of 26)
Overall | |
Schiller1 | 100.0000 |
Citology1 | 95.9648 |
Biopsy1 | 61.0357 |
Hinselmann1 | 53.8580 |
Dx.Cancer | 4.4652 |
Age | 1.9525 |
Dx | 1.6306 |
Dx.CIN | 1.5675 |
First.sexual.intercourse | 0.4283 |
Remaining attributes were shown with 0.0 value. The significance of variables in rpart method is shown with the following plot in Fig. 9.
C5.0()—The C50 package in R contains an interface to the C5.0 model. This method acknowledges noise and missing values in the dataset. This method can appropriately anticipate relevant attributes in the dataset, the problem of overfitting and error pruning is solved with this algorithm [21]. The plot for variable importance through C5.0 method is shown below in Fig. 10.
rf()—Random forests are based on decision trees. They also have feature importance methodology which uses ‘gini index’ to assign a score and rank the features based on the values [22, 23]. The following plot in Fig. 11 shows the variable importance by applying rf method.
ctree()—Conditional inference trees evaluate a regression association by binary recursive partitioning in a conditional inference framework. ctree uses a significant procedure to select variables. Ctree is based on a overall theory of permutation tests, executing a hypothesis test at each node, accordingly producing a equivalent p value to test whether the tree should stop or keep growing [24]. The following plot in Fig. 12 shows the variable importance by means of ctree method.
SVM and KNN—The principle of SVM classifier (Support Vector Machine) method is to build a hyperplane separating data for different classes. The main consideration while drawing the hyperplane is on maximizing the distance from hyperplane to the nearest data point of either class. These adjacent data points are known as Support Vectors. KNN algorithm is an instance-based learning algorithm which calculates distance for a particular value of K for each new sample. SVM and KNN methods showed the ROC curve variable importance. In both the methods variables are sorted by maximum importance across the classes.
4.2.5 Determining significant features
Through our experiments with various feature selection methods, it is observed that the features which are most important in few algorithms are not equally important in other algorithms. This work is proposed to decide about the optimal number of features and also to choose most significant features from all these procedures. The proportional study of these outcomes attained through various feature selection methods have shown that there are few features which are significant in all the methods other than those four target variables. Similarly, some features are common or showing more percentage of importance in few methods. Hence it is decided to select the significant predictors based on the higher rank (percentage) value gained and based on their mutual existence in a greater number of feature selection methods. As we combined four target features Hinselmann, Schiller, Citology and Biopsy as a single target variable, they could be considered as most significant features. Apart from this based on the proportion of importance (ranks), commonality and precedence in finding the exact outcome, additional ten core features have been identified as most significant to predict the final result ‘Cancer’. They are mentioned below.
Hormonal.Contraceptives..years | Dx.Cancer |
First.sexual.intercourse | Dx |
Number.of.sexual.partners | Dx.HPV |
STDs..Number.of.diagnosis | Smokes..years. |
Age | Num.of.pregnancies |
It is observed through Fig. 12 which shows the significance of variables through ctree() method has selected most of these significant features in an efficient manner.
4.3 Classifier model construction and estimation of model accuracy
Feature Selection through ML algorithms have already trained the desired models for classification. Once the best feature selection subset is identified for a particular dataset the same can be used to improve the classifier accuracy [18]. Hence, to enhance the performance and accuracy of various classifier models, decided to apply boosting method in determining the occurrence in cervical cancer [25]. When we are building a predictive model there must be a way to evaluate the capability of the model on concealed data. This can be accomplished by estimating accuracy through the data that was not used to train the model such as test data or by means of cross validation. The model should be trained on a large percentage of the dataset. Correspondingly there is a necessity for a good ratio of testing data points, because fewer amount of data points can lead to a variance error while testing the model effectiveness. It is essential that training and testing process should be iterated multiple times, correspondingly the training and testing dataset distribution should be changed which helps to accurately validate the effectiveness of the model. All these requirements could be attained through K-fold cross validation.
4.3.1 K-fold cross validation
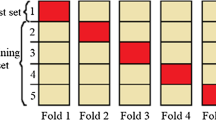
The K-fold cross validation method comprises splitting the dataset into k-subsets, where each subset/fold is used as a testing set. In the first iteration, first fold is used for model testing and the rest are used for model training. Likewise, this process will be repeated until each fold have been used for testing the model. The picturing of k-fold cross validation with k = 10 is shown in Fig. 13. This method is useful in defining the accuracy of the model with reasonable combinations of data.
4.3.2 Repeated k-fold cross validation
The procedure of splitting the data into k-folds can be repeated for a required number of times, which is known as Repeated k-fold Cross Validation. The eventual model accuracy is calculated as the mean from the number of repeats. In this work, repeated cross validation techniques have been applied for the processes of data splitting, model training and testing in recurrent manner for a greater (for 50) number of times over cervical cancer data. The trainControl() function in R can be used to specify resampling type. The code to apply repeated cross validation for fifty times is shown here.
control <- trainControl(## 10-fold CV |
method = “repeatedcv”, |
number = 10, |
## repeated fifty times repeats = 50) |
The train() function in R is used to fit the predictive models based on various tuning parameters. The model training thorough Random Forest technique is shown below.
-
fit.rf < - train(Cancer ~ ., data = cer_data, method = ”rf”, trControl = control)
The results attained by rf method using 26 predictor (features) values with 10-fold cross validation through 50 repeats are shown below.
Random Forest
-
600 samples
-
26 predictor
-
5 classes ‘0’, ‘1’, ‘2’, ‘3’, ‘4’
-
No pre-processing
-
Resampling Cross-Validated (10 fold, repeated 50 times)
-
Summary of sample sizes 541, 540, 541, 541, 540, 540,…
-
Resampling results across tuning parameters:
mtry | Accuracy | Kappa |
2 | 0.9111862 | 0.4565576 |
14 | 0.9879466 | 0.9445185 |
26 | 0.9908683 | 0.9578242 |
-
Accuracy was used to select the optimal model using
-
the largest value.
-
The final value used for the model was mtry = 26.
The accuracy of rf model with repeated cross validation using 26 features is revealed through the plot as shown in Fig. 14.
The results attained by rf method using 14 predictor (features) values with 10-fold cross validation with 50 repeats are shown below.
Random Forest
-
600 samples
-
14 predictor
-
5 classes ‘0’, ‘1’, ‘2’, ‘3’, ‘4’
-
No pre-processing
-
Resampling Cross-Validated (10 fold, repeated 50 times)
-
Summary of sample sizes 540, 539, 539, 540, 540, 540,…
-
Resampling results across tuning parameters:
mtry | Accuracy | Kappa |
2 | 0.9597476 | 0.8016736 |
8 | 0.9901440 | 0.9548854 |
14 | 0.9927090 | 0.9669582 |
-
Accuracy was used to select the optimal model using
-
the largest value.
-
The final value used for the model was mtry = 14.
Similarly, the accuracy plot of rf model with repeated cross validation using 14 features is shown in Fig. 15.
Correspondingly, additional classifier models have been created with maximum possibilities on this cervical cancer data through various methods like rpart, C5.0, SVM and KNN over repeated k-fold cross validation by 50 trials to determine whether the results obtained are significant in other algorithms as well. These results are revealed in Fig. 16 by printing and plotting some of these model outputs.
The results attained are comparatively upgraded for most of the ML methods by training the models with these significant features (14 Predictors) which were obtained through feature selection processes. The accuracy attained for SVM model has shown very good progression with significant features but KNN model has not accomplished progressive results. The plot for these models is shown in Fig. 17.
This exploration proved that the results attained are more significant by applying repeated cross validation techniques for training the models. We have selected ten significant features which are optimal with the percentage (rank) values obtained and also based on their commonality existence in a greater number of feature selection methods, subsequently the results are more precise and significant.
4.4 Performance and accuracy estimation of ML classifier models
To enhance the efficiency of clinical outcome predictions multiple measurements can be used as performance metrics [26]. In this study the performance competences of various classification methods are measured using the evaluation metrics like accuracy and AUC (Area Under Curve) values. Accuracy is one of the metrics for assessing classification models which is calculated as the fraction of predictions our model got precise and the formula is given below.
In this work, accuracy is estimated for some of the prevalent classification algorithms like C5.0, rpart, rf, SVM and KNN in two ways i.e. by considering 26 input features in the dataset (without feature selection process) and by considering the significant features (14 features) obtained through feature selection methods which are discussed earlier. The results attained for these classifiers are revealed through cross tables, performance accuracy and AUC values.
4.4.1 Accuracy of ML models with 26 features
The accuracy obtained for conferred ML models with 26 predictors are exhibited through confusion matrix with the actual and predicted outcomes.
The confusion matrix in Fig. 18 shows the output of C5.0 procedure, and this output is obtained by using 26 features of the dataset.
-
Performance of C5.0 algorithm: Accuracy = 97%, AUC = 0.91
The confusion matrix in Fig. 19 shows the output of rpart method with 26 features.
-
Performance of rpart algorithm: Accuracy = 96%, AUC = 0.81
The confusion matrix output of rf algorithm is shown in Fig. 20.
-
Performance of rf algorithm: Accuracy = 96.9%, AUC = 0.91
Likewise, the outcomes were accomplished for SVM and KNN models also. But their accuracy (88%) and AUC (0.5) values are comparatively less than other methods. The precisions obtained for these ML models are displayed through Box and Whisker Plots in Fig. 21 which is a suitable way to look at the spread of the estimated accuracies for varied methods and how they relate.
4.4.2 Accuracy of ML models with feature selection process
The accuracy obtained for conferred ML models with 14 predictors are revealed through confusion matrix based on the actual and predicted results.
Output of C5.0 and rf Methods with Selected Features:
The implementation of C5.0 and rf algorithms on cervical cancer dataset with selected features have shown highest accuracy (100%) with AUC as 0.91. The evaluation on training data is shown below in Fig. 22.
The confusion matrix output of C5.0 and rf methods are depicted in Fig. 23. This model is extremely accurate at 99.77%.
-
Performance of C5.0 and rf algorithms: Accuracy = 100%, AUC = 0.91
Similarly, the output for rpart algorithm has attained 97% accuracy with 0.81 as AUC and SVM method attained 93% accuracy with 0.8 as AUC. But the performance of KNN has not much upgraded, it has shown 89% as accuracy and 0.5 as AUC. The enhancement in the precision values of these classifiers with an optimal feature subset is shown in Fig. 24.
The results showed that C5.0 and rf methods equally attained well with maximum accurateness and SVM model has achieved a better improvement with optimal features.
5 Results and discussion
In this work, Machine Learning algorithms (C5.0, RF, RPART, SVM and KNN) were employed for cervical cancer diagnosis to prove the importance of model building with data cleaning, replacement of missing values and applying feature selection process to achieve higher efficiency in outcome prediction with an optimal feature subset. To evaluate the performance of classifier models, this work employed ML methods on the cervical cancer data by considering all the records in the dataset through replacement of missing values in the rows with their mean, eliminating only the columns which had missing values. Hence, after data cleaning process the dataset had 858 rows with 26 predictors. Then by implementing few imperative feature selection techniques and by training the models through ML algorithms, an optimal feature subset has been selected based on the importance of variables. The following attributes have been identified as more significant in addition with four target features for cervical cancer diagnosis prediction.
Hormonal.Contraceptives..years | Dx.Cancer |
First.sexual.intercourse | Dx |
Number.of.sexual.partners | Dx.HPV |
STDs..Number.of.diagnosis | Smokes..years. |
Age | Num.of.pregnancies |
ML classifier models with C5.0, RF, RPART, SVM and KNN methods have been built with repeated k-fold cross validation technique with all the 26 features as well as with an optimal feature subset of 14 predictors for diagnosis prediction of cervical cancer. The results of the classifier models through C5.0 and rf algorithms with an optimal significant feature are significantly upgraded to 99% to 100%. In both the ways this work revealed that C5.0 and rf methods as more prominent algorithms for predicting significant risk factors in cervical cancer. The relative performance analysis of the conferred classification methods is shown in Table 2 with their accuracy and AUC values.
The performance evaluation of ML classification algorithms is exhibited through the bar plot which is shown in Fig. 25. Random forest and C5.0 both the methods have equally performed well with maximum accuracy and reduced amount of error rate.
We have selected significant predictors based on their importance and mutual existence over feature selection methods and by training the models through repeated k-fold cross validation with ML methods. Through unbiased feature list, there are only three features which are common in most of these methods. If we employ these minimal features for ML classification process then the results will not be precise. This shows that the stability in feature selection is an important issue and its importance has been determined through this work. Therefore, an optimized feature selection approach is more essential to improve the performance accuracy of prediction process, accordingly with an optimal feature subset an efficient performance has been gained through this work for cervical cancer diagnosis prediction.
6 Conclusion
Cervical cancer is one of the important reasons among female cancer deaths in the recent years. But, through machine learning, we are able to recognize the factors that increase possibility of evolving this cancer in women. The feature selection process over Boruta algorithm, SA and ctree() methods have shown good proficiency in accomplishing major features of an optimal feature subset for cervical cancer risk factors prediction. However, all the information related to the dataset were not provided and some of the information, such as factorizing or not factorizing, replacement of variables was done based on assumptions. Through the examination of C5.0, rpart, Random Forest, SVM and KNN algorithms, we have found that most of the algorithms were efficient in providing cervical cancer diagnosis with advanced accuracy. Overall C5.0 and Random Forest classifiers have performed reasonably well, besides extremely accurate through reliable results with maximum accuracy for identifying women exhibiting clinical sign of cervical cancer. It is apparent through this work that, an enhanced prediction accuracy for cervical cancer diagnosis can be attained by means of including an optimal feature subset through enhanced feature selection approaches and by building the classifier models with ML algorithms through repeated k-fold cross validation techniques. This work can be extended for other types of gynecological cancer type predictions. Altogether, the conferred classifiers have shown enhanced performance accuracy with the optimal features’ dataset.
References
World Health Organization (2019) Fact sheet: human-papillomavirus-(hpv)-and-cervical-cancer, Retrieved 13-02-2019
Sarwar A et al (2015) Performance evaluation of machine learning techniques for screening of cervical cancer, INDIACom-2015; ISSN 0973-7529; ISBN 978-93-80544-14-4
Abdoh SF, Abo Rizka M, Maghraby FA (2018) Cervical cancer diagnosis using random forest classifier with SMOTE and feature reduction techniques. In: IEEE Access, vol 6, pp 59475–59485
Kourou K et al (2015) Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J 13:8–17
Bischl B et al (2016) mlr: machine learning in R. J Mach Learn Res 17:1–5
Gowda A et al (2010) Feature subset selection problem using wrapper approach in supervised learning. Int J Comput Appl 1(7):13–17
Lavanya D et al (2011) Analysis of feature selection with classification: breast cancer datasets. Indian J Comput Sci Eng (IJCSE) 2(5):756–763
Sowjanya D et al (2014) Staging prediction in cervical cancer patients—a machine learning approach. Int J Innov Res Pract 2(2):14–23
Akyol K (2018) A study on test variable selection and balanced data for cervical cancer disease. Int J Inf Eng Electron Bus 10:1
Menon V, Parikh D (2018) Machine learning applied to cervical cancer data. Int J Sci Eng Res 9(7):46–50
Choudhary A et al (2018) Classification of cervical cancer dataset. In: Proceedings of the 2018 IISE annual conference, Orlando, pp 1456–1461
Jović A, Brkić K, Bogunović N (2015) A review of feature selection methods with applications. In: 2015 38th international convention on information and communication technology, electronics and microelectronics (MIPRO), Opatija, pp 1200–1205
Bagherzadeh-Khiabani F et al (2016) A tutorial on variable selection for clinical prediction models: feature selection methods in data mining could improve the results. J Clin Epidemiol 71:76–85
Le Thi HA et al (2015) Feature selection in machine learning: an exact penalty approach using a difference of convex function algorithm. Mach Learn 101:163–186
Park HW et al (2017) A hybrid feature selection method to classification and its application in hypertension diagnosis. In: ITBAM 2017, LNCS 10443. Springer, pp 11–19
Ruiz R et al (2006) Incremental wrapper-based gene selection from microarray data for cancer classification. Pattern Recognition 39(12):2383–2392
UCI Machine Learning Repository, Cervical cancer (Risk Factors) Data Set. Retrieved February 5, 2019, from https://archive.ics.uci.edu/ml/datasets/Cervical+cancer+%28Risk+Factors%29
Zhao Z et al (2010) Advancing feature selection research—ASU feature selection repository: Citeseer
Rudnicki WR, Wrzesień M, Paja W (2015) All relevant feature selection methods and applications. In: Stańczyk U, Jain L (eds) Feature selection for data and pattern recognition. Studies in computational intelligence, vol 584. Springer, Berlin
Antony DA (2016) Literature review on feature selection methods for high-dimensional data. Int J Comput Appl 136:0975–8887
Pandya R, Pandya J (2015) C5.0 algorithm to improved decision tree with feature selection and reduced error pruning. Int J Comput Appl 117(16):18–21
Nguyen C et al (2013) Random forest classifier combined with feature selection for breast cancer diagnosis and prognostic. J Biomed Sci Eng 6:551–560
Genuer R et al (2015) An R package for variable selection using random forests. The R J R Found Stat Comput 7(2):19–33
Jacobucci R (2018) Decision tree stability and its effect on interpretation. Retrieved from osf.io/m5p2v
Dinov ID (2018) Improving model performance. In: Data science and predictive analytics. Springer, Cham, pp 497–511
Seethal CR, Panicker JR, Vasudevan V (2016) Feature selection in clinical data processing for classification. In: International conference on information science (ICIS), pp 172–175
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nithya, B., Ilango, V. Evaluation of machine learning based optimized feature selection approaches and classification methods for cervical cancer prediction. SN Appl. Sci. 1, 641 (2019). https://doi.org/10.1007/s42452-019-0645-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-019-0645-7