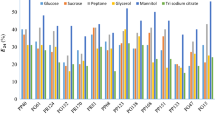
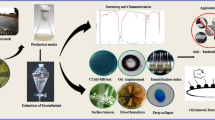
Abstract
Biosurfactants are low molecular weight, surface active amphiphatic compounds produced by microorganisms. These molecules could be of wide acceptability in cosmetic, pharmaceutical, food industries and bioremediation processes due to low toxicity and biodegradability. However, there is a limitation in their applicability partially due to our knowledge that revolves around pretty less number of well studied biosurfactants. The aim of the present study is to isolate bacteria from oil contaminated environments (soil from automobile workshops and kitchen waste dumping area), and evaluate them for biosurfactant production and determining the properties of these biomolecules. Isolates were screened for drop collapse activity, emulsification, phenol–sulfuric test and ability of adhesion to hydrocarbons. Isolates selected on the basis of above activities were screened for antimicrobial activity against Bacillus cereus, Escherichia coli, Klebsiella sp., Pseudomonas aeruginosa and Staphylococcus aureus. Biosurfactants with antimicrobial activity were also checked for ability to reduce adhesion of pathogenic microorganisms to solid surface and evaluated for biofilm-disruption activity. The results obtained in this work showed that the biosurfactant from isolate SBM1 identified as Bacillus sp. had all the above properties. Thin layer chromatography (TLC) and Fourier transform infrared spectroscopy (FTIR) analysis confirmed that biosurfactant from isolate SBM1 is of glycolipid type that can be employed as potential agent of biomedical utility.


Similar content being viewed by others
References
Altschul SF, Madden TL, Scaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search program. Nucleic Acids Res 25:3389–3402
Ariech M, Guechi A (2015) Assessment of four different methods for selecting biosurfactant producing extremely halophilic bacteria. African J Biotechnol 14(21):1764–1772
Banat IM, Franzetti A, Gandolfi I, Bestetti G, Martinotti MG, Fracchia L, Smyth TJ, Marchant R (2010) Microbial biosurfactants production, applications and future potential. Appl Microbiol Biotechnol 87:427–444
Bodour A, Drees K, Maier R (2003) Distribution of biosurfactant-producing bacteria in undisturbed and contaminated arid southwestern soils. Appl Environ Microbiol 69(6):3280–3287
Carniello V, Peterson BW, van der Mei HC, Busscher HJ (2018) Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth. Adv Colloid Interfac 261:1–14
Cooper DG, Goidenberg BG (1987) Surface-active agents from two Bacillus Species. Appl Environ Microbiol 53(2):224–229
Das P, Mukherjee S, Sen R (2008) Antimicrobial potential of a lipopeptide biosurfactant derived from a marine Bacillus circulans. J Appl Microbiol 104:1675–1684
Das P, Mukherjee S, Sen R (2009) Antiadhesive action of a marine microbial surfactant. Colloid Surface B 71:183–186
Díaz De Rienzo MA, Banat IM, Dolman B, Winterburn J, Martin PJ (2015) Sophorolipid biosurfactants: possible uses as antibacterial and antibiofilm agent. N Biotechnol 32:720–726
Dubois M, Gills KA, Hailton JK, Reberes PA, Smit F (1956) Colorimetric method for determination of sugar and realted substances. Anal Chem 28:350–356
Dusane DH, Nancharaiah V, Zinjarde SS, Venugopalan VP (2010) Rhamnolipid mediated disruption of marine Bacillus pumilus biofilms. Colloid Surface B 81:242–248
Elshikh M, Marchant R, Banat IM (2016) Biosurfactants: promising bioactive molecules for oral-related health applications. FEMS Microbiol Lett 363(18):fnw213
Garrity G (2005) Bergey’s manual of systematic bacteriology. In: Brenner DJ, Krieg NR, Staley JT (eds) The proteobacteria, Part B. The gammaproteobacteria, vol. 2, 2nd edn. Springer, New York, pp 323–379
Giaouris E, Chapot-Chartier M-P, Briandet R (2009) Surface physicochemical analysis of natural Lactococcus lactis strains reveals the existence of hydrophobic and low charged strains with altered adhesive properties. Int J Food Microbiol 131:2–9
Górna H, Ławniczak Ł, Zgoła-Grześkowiak A, Kaczorek E (2011) Differences and dynamic changes in the cell surface properties of three Pseudomonas aeruginosa strains isolated from petroleum-polluted soil as a response to various carbon sources and the external addition of rhamnolipids. Bioresour Technol 102(3):3028–3033
Govindammal M, Parthasarathi R (2013) Investigation on antimicrobial activity of biosurfactnt produced by Pseudomonas fluorescens isolated from mangrove ecosystem. Int Res J Pharm 4(1):230–232
Gudiña EJ, Rangarajan V, Sen R, Rodrigues LR (2013) Potential therapeutic applications of biosurfactants. Trends Pharmacol Sci 34:667–675
Gudiña EJ, Fernandes EC, Rodrigues AI, Teixeira JA, Rodrigues LR (2015) Biosurfactant production by Bacillus subtilis using corn steep liquor as culture medium. Front Microbiol 6:59. https://doi.org/10.3389/fmicb.2015.00059
Günzler H, Gremlich HU (2003) IR-spektroskopie, 1st edn. Wiley-VCH, Weinheim
Jain D, Collins-Thompson D, Lee H et al (1991) A drop-collapsing test for screening surfactant-producing microorganisms. J Microbiol Methods 13(4):271–279
Janek T, Krasowska A, Czy˙znikowska Z, Łukaszewicz M (2018) Trehalose lipid biosurfactant reduces adhesion of microbial pathogens to polystyrene and silicone surfaces: an experimental and computational approach. Front Microbiol 9:2441
Khire JM (2010) Bacterial biosrufactants, and their ole in microbial enhanced oil recovery (MEOR). In: Sen R (ed) Biosurfactants. Advances in experimental medicine and biology, vol 672. Springer, New York
Kuyukina MS, Ivshina IB, Korshunova IO, Stukova GI, Krivoruchko AV (2016) Diverse effects of a biosurfactant from Rhodococcus ruber IEGM 231 on the adhesion of resting and growing bacteria to polystyrene. AMB Express 6(1):14
Liu G, Fang Z, Zhong H, Shi L, Yang X, Liu Z (2020) Transport of Pseudomonas aeruginosa in porous media mediated by low-concentration surfactants: The critical role of surfactant to change cell surface hydrophobicity. Water Resour Res 56:e2019WR026103
Maidak BL, Olsen GL, Larsen N, Overbeek R, McCaughey MJ, Woese CR (1997) The ribosomal database project. Nucleic Acids Res 24:82–85
Maidak BL, Cole JR, Lilburn TG, Parker CT, Saxman PR, Stredwick JM, Garrity GM, Li B, Olsen GL, Pramanik S, Schmidt TM, Tiedje JM (2000) The RDP (Ribosomal Database Project) continues. Nucleic Acids Res 28:173–174
Mangwani N, Kumari S, Das S (2016) Bacterial biofilms and quorum sensing: fidelity in bioremediation technology. Biotechnol Genet Eng 32(1–2):43–73
Mani P, Dineshkumar G, Jayaseelan T, Deepalakshmi K, Kumar G, Senthil C, Balan S (2016) Antimicrobial activities of a promising glycolipid biosurfactant from a novel marine Staphylococcus saprophyticus SBPS 15. 3 Biotech 6(2):163
Moshtagh B, Hawboldt K, Zhang B (2019) Optimization of biosurfactant production by Bacillus subtilis N3–1P using the brewery waste as the carbon source. Environ Technol 40(25):3371–3380
Nayarisseri A, Singh P, Singh SK (2018) Screening, isolation and characterization of biosurfactant producing Bacillus subtilis strain ANSKLAB03. Bioinformation 14(6):304–314
Nitschke M, Pastore GM (2004) Biosurfactant production by Bacillus subtilis using cassava-processing effluent. Appl Biochem Biotechnol 112(3):163–172
Ocampo GY (2016) Role of biosurfactants in nature and biotechnological applications. J Bacteriol Mycol 2(4):95–96
Pacwa-Plociniczak M, Plaza GA, Piotrowska-Seget Z, Cameotra SS (2011) Environmental applications of biosurfactants: recent advances. Int J Mol Sci 12:633–654
Patowary K, Patowary R, Kalita MC, Deka S (2017) Characterization of biosurfactant produced during degradation of hydrocarbons using crude oil as sole source of carbon. Front Microbiol 8:279. https://doi.org/10.3389/fmicb.2017.00279
Rani M, Weadge JT, Jabaji S (2020) Isolation and characterization of biosurfactant-producing bacteria from oil well batteries with antimicrobial activities against food-borne and plant pathogens. Front Microbiol 11:64
Rosenberg M, Gutnick D, Rosenberg E (1980) Adherence of bacteria to hydrocarbons—a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett 9(1):29–33
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Harbor Laboratory Press, Cold Spring Harbor, New York
Santos D, Rufino R, Luna J, Santos V, Sarubbo L (2016) Biosurfactants: multifunctional biomolecules of the 21st century. Int J Mol Sci 17(3):401
Satpute S, Mone N, Das P, Banpurkar A, Banat I (2018) Lactobacillus acidophilus derived biosurfactant as a biofilm inhibitor: A promising investigation using microfluidic approach. Appl Sci 8(9):555
Satpute SK, Mone NS, Das P, Banat IM, Banpurkar AG (2019) Inhibition of pathogenic bacterial biofilms on PDMS based implants by L. acidophilus derived biosurfactant. BMC Microbiol 19(1):39. https://doi.org/10.1186/s12866-019-1412-z
Shah S, Bhattarai A, Chatterjee S (2011) Surfactants, its applications and effects on environment. Bibechana 7:61–64. https://doi.org/10.3126/bibechana.v7i0.4047
Shao B, Liu Z, Zhong H, Zeng G, Liu G, Yu M, Liu Y, Yang X, Li Z, Fang Z, Zhang J, Zhao C (2017) Effects of rhamnolipids on microorganism characteristics and applications in composting: a review. Microbiol Res 200:33–44
Sidkey NM, Mohamed HF, Elkhouly HI (2016) Evaluation of different screening methods for biosurfactant producers isolated from contaminated Egyptian samples grown on industrial olive oil processing waste. Brit Microbiol Res J 17(4):1–19. https://doi.org/10.9734/BMRJ/2016/28437
Silva SNRL, Farias CBB, Rufino RD, Luna JM, Sarubbo L (2010) Glycerol as substrate for the production of biosurfactant by Pseudomonas aeruginosa UCP0992. Colloid Surface B 79:174–183
Silva J, Maria N, Rocha P, Rufino RD, Luna JM, Silva RO, Sarubbo LA (2014) Characterization of a biosurfactant produced by Pseudomonas cepacia CCT6659 in the presence of industrial wastes and its application in the biodegradation of hydrophobic compounds in soil. Colloid Surface B 117:36–41
Singh P, Tiwary BN (2016) Isolation and characterization of glycolipid biosurfactant produced by a Pseudomonas otitidis strain isolated from Chirimiri coal mines. India Bioresour Bioprocess 3:42
Souza EC, Vessoni Penna T, Pinheiro De Souza R (2014) Biosurfactant–enhanced hydrocarbon bioremediation: an overview. Int Biodeterior Biodegrad 89:88–94
Tahzibi A, Kamal F, Assadi MM (2004) Improved production of rhamnolipids by a Pseudomonas aeruginosa mutant. Iran Biomed J 8(1):25–31
Talukdar P, Sharma C, Doley A, Baruah K, Borah A, Agarwal P, Deori P (2017) Isolation and characterization of biosurfactant producing microorganisms from petroleum contaminated soil samples for EOR and bioremediation. Petrol Sci Technol 35(22):2102–2108
Thavasi R, Jayalakshmi S, Balasubramanian T, Banat IM (2007) Production and characterization of a glycolipid biosurfactant from Bacillus megaterium using economically cheaper sources. World J Microbiol Biotechnol 24(7):917–925
Thavasi R, Sharma S, Jayalakshmi S (2011) Evaluation of screening methods for the isolation of biosurfactant producing marine bacteria. J Pet Environ Biotechnol S1:001. https://doi.org/10.4172/2157-7463.S1-001
Turkovskaya OV, Dmitrieva TV, Muratova AY (2001) A biosurfactant-producing Pseudomonas aeruginosa strain. Appl Biochem Microbiol 37(1):71–75
Yuan X, Ren F, Zeng G, Zhong H, Fu H, Liu J, Xu X (2007) Adsorption of surfactants on a Pseudomonas aeruginosa strain and the effect on cell surface lypohydrophilic property. Appl Microbiol Biotechnol 76(5):1189–1198
Yuliani H, Perdani MS, Savitri I, Manurung M, Sahlan M, Wijanarko A, Hermansyah H (2018) Antimicrobial activity of biosurfactant derived from Bacillus subtilis C19. Energy Proc 153:274–278
Zezzi do Valle Gomes N, Nitschke N (2012) Evaluation of rhamnolipid and surfactin to reduce the adhesion and remove biofilms of individual and mixed cultures of food pathogenic bacteria. Food Control 25(2):441–447
Acknowledgements
The authors Khare and Verma are grateful to Vice Chancellor, Chhatrapati Shahu Ji Maharaj University, Kanpur, India for providing facilities.
Author information
Authors and Affiliations
Contributions
EK: responsible for work idea, experimentation and manuscript preparation; EV: Performed experiments.
Corresponding author
Ethics declarations
Consent to participate
Both the authors are involved in experimentation. Manuscript written by Dr. Ekta Khare.
Consent for publication
Both the authors give consent for the publication of manuscript in “Environmental Sustainability” and it is not been submitted anywhere else.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
42398_2020_136_MOESM1_ESM.ppt
Supplementary Figure S1. Gel electrophoresis shows the ~1.5 kb amplified fragment of 16S rRNA gene from isolate SBM1. Where, M= 100 bp-10 kb wide range DNA marker
42398_2020_136_MOESM2_ESM.ppt
Supplementary Figure S2. Universal primers 27F and 1492R used for the amplification of ~1.5 kb 16S rRNA gene sequence of isolate SBM1
42398_2020_136_MOESM3_ESM.ppt
Supplementary Figure S3. Sequence of ~1.5 kb 16S rRNA gene sequence from Bacillus strain SBM1 submitted to NCBI GenBank under accession number MN428789
Rights and permissions
About this article
Cite this article
Khare, E., Verma, E. Bacteria from oil contaminated sites as a viable source of potential biosurfactants with anti-microbial and anti-biofilm activities. Environmental Sustainability 3, 497–507 (2020). https://doi.org/10.1007/s42398-020-00136-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42398-020-00136-8