Abstract
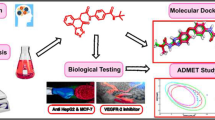
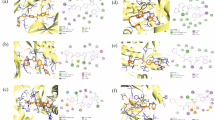
Cancer is alleged to be the second major mortality inflicting malady according to the world health organization (WHO) reports. Even though there are high advances in the diagnosis and remedy of this ailment, the patient’s survival ratio remains unsatisfactory as a result of the prevalent antagonistic effects of anticancer drugs. Hence, the discovery of new, effective, selective, and much less toxic anticancer agents is still mandatory. In this study we report the design of new 3-methylquinoxaline derivatives for the inhibition of VEGFR-2 receptors through molecular docking and structural modification of the template compound. The results of docking study performed between forty one series of 3-methylquinoxaline derivatives and the active site of the VEGFR-2 target receptor (pdb id = 2OH4) illustrates that five compounds (5, 27, 33, 37 and 40) showed higher MolDock and Rerank scores than Sorafenib (control drug), as such they were tagged as the potential hit compounds. Compound 5 having the highest docking scores (MolDock score = -156.063, Rerank score = -133.13) with good pharmacokinetic profile, and drug-likeness properties was utilized as a template for the design of eight new novel 3-methyl quinoxaline derivatives. All the designed compounds were found to have better docking scores ranging from − 156.966 to -167.961 Moldock scores and − 133.485 to -137.852 Rerank scores compared to the template compound (Moldock score = -156.063, Rerank score = -133.13) and Sorafenib (Moldock score = -146.535, Rerank score = -102.625). The designed compounds do not violate the acceptable criteria set by Lipinski, hence, they are easily transported, absorbed and diffused in to the human system. Additionally, they have low predicted synthetic accessibility values ranging from 3.29 to 3.85 indicating that they are easily synthesized in the laboratory and high bioavailability scores of 0.55 which revealed that they are orally bioavailable. Hence, the designed 3-methylquinoxaline derivatives can serve as better VEGFR-2 inhibitors.










Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I et al (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424
Barbosa IR, Souza D, LB d, Bernal MM (2015) Costa _I. d CC. Cancer mortality in Brazil: temporal trends and predictions for the year 2030. Medicine 94:e746
El Newahie AM, Ismail NS, Abou El Ella DA, Abouzid KA (2016) Quinoxaline-based scaffolds targeting tyrosine kinases and their potential anticancer activity. Arch Pharm 349:309–326
Chu H, Wang Y (2012) Therapeutic angiogenesis: controlled delivery of angiogenic factors. Ther Deliv 3:693–714
Sajib S, Zahra FT, Lionakis MS et al (2018) Mechanisms of angiogenesis in microbe-regulated inflammatory and neoplastic conditions. Angiogenesis 21:1–14
Guo S, Colbert LS, Fuller M et al (2010) Vascular endothelial growth factor receptor-2 in breast cancer. Biochim Biophys Acta Rev Cancer 1806:108–121
Takahashi S (2011) Vascular endothelial growth factor (VEGF), VEGF receptors and their inhibitors for antiangiogenic tumor therapy. Biol Pharm Bull 34:1785–1788
Holmes K, Roberts OL, Thomas AM, Cross MJ (2007) Vascular endothelial growth factor receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell Signal 19:2003–2012
Hamza AS, Uzairu A, Shallangwa GA, Sani Uba & Abdullahi Bello Umar (2022) Molecular Docking, ADMET and Pharmacokinetic properties predictions of some di-aryl Pyridinamine derivatives as Estrogen Receptor (Er +) Kinase Inhibitors. Egypt J Basic Appl Sci 9(1):180–204. DOI: https://doi.org/10.1080/2314808X.2022.2050115
Xu F, Cheng G, Hao H, Wang Y, Wang X, Chen D, Peng D, Liu Z, Yuan Z, Dai M (2016) Mechanisms of Antibacterial Action of Quinoxaline 1,4-di-N-oxides against Clostridium perfringens and Brachyspira hyodysenteriae. Front Microbiol. ;7:1948. doi: https://doi.org/10.3389/fmicb.2016.01948
Mónica Vieira C, Pinheiro (2014) Rúben Fernandes, João Paulo Noronha, Cristina Prudêncio, Antimicrobial activity of quinoxaline 1,4-dioxide with 2- and 3-substituted derivatives. Microbiol Res 169(4):287–293
Tang X, Zhou Q, Zhan W, Sun DHuRZhouN, Chen S (2022) Wenneng Wu and Wei Xue. Synthesis of novel antibacterial and antifungal quinoxaline derivatives. RSC Adv 12:2399
Asuncion Burguete E, Pontiki D, Hadjipavlou-Litina S, Ancizu R, Villar B, Solano E, Moreno E, Torres (2011) Silvia Perez, Ignacio Aldana and Antonio Monge. Synthesis and Biological Evaluation of New Quinoxaline Derivatives as Antioxidant and Anti-Inflammatory Agents. Chem Biol Drug Des 77:255–267
Mohammed M, Alanazi A, Elwan NA, Alsaif AJ, Obaidullah, Hamad M, Alkahtani, Abdulrahman A, Al-Mehizia SM, Alsubaie, Mohammed S, Taghour, Ibrahim H, Eissa (2021) Discovery of new 3-methylquinoxalines as potential anti-cancer agents and apoptosis inducers targeting VEGFR-2: design, synthesis, and in-silico studies. J Enzyme Inhib Med Chem 36(1):1732–1750. DOI: https://doi.org/10.1080/14756366.2021.1945591
Mohammed M, Alanazi IH, Eissa NA, Alsaif, Ahmad J, Obaidullah WA, Alanazi AF, Alasmari H, Albassam H, Elkady, Alaa Elwan (2021) Design, synthesis, docking, ADMET studies, and anticancer evaluation of new 3- methylquinoxaline derivatives as VEGFR-2 inhibitors and apoptosis inducers. J Enzyme Inhib Med Chem 36(1):1760–1782. DOI: https://doi.org/10.1080/14756366.2021.1956488
Macalino SJ, Billones JB, Organo VG, Carrillo MC (2020) In-silico strategies in tuberculosis drug discovery. Molecules 25(3):665
Anusuya S, Velmurugan D, Gromiha MM (2016) Identification of dengue viral RNA-dependent RNA polymerase inhibitor using computational fragment-based approaches and molecular dynamics study. J Biomol Struct Dyn 34(7):1512–1532
Benmansour F, Eydoux C, Querat G, de Lamballerie X, Canard B, Alvarez K, Guillemot JC, Barral K (2016) Novel 2-phenyl-5-[(E)-2-(thiophen-2-yl) ethenyl]-1, 3, 4-oxadiazole and 3-phenyl-5-[(E)-2-(thiophen-2-yl) ethenyl]– 1, 2, 4-oxadiazole derivatives as dengue virus inhibitors targeting NS5 polymerase. Eur J Med Chem 109:146–156
Meng XY, Zhang HX, Mezei M, Cui M (2011) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 7(2):146–157
Hamza AS, Uzairu A, Shallangwa GA, Sani Uba and Abdullahi Bello Umar (2021) In–silico activity prediction, structure–based drug design, molecular docking and pharmacokinetic studies of selected quinazoline derivatives for their antiproliferative activity against triple negative breast cancer (MDA–MB231) cell line
Hamza AS, Ibrahim UA et al (2021) Tukur Ibrahim and Abdullahi Bello Umar. Chemoinformatics activity prediction, ligand based drug design, Molecular docking and Pharmacokinetics studies of some series of 4, 6diaryl2pyrimidinamine derivatives as anticancer agents. Bull Natl Res Cent 45(1):167
Masaichi Hasegawa N, Nishigaki Y, Washio et al (2007) Discovery of novel Benzimidazoles as potent inhibitors of TIE-2 and VEGFR-2 Tyrosine Kinase Receptors. J Med. Chem; 50: 4453–5570
Umar BA, Uzairu A, Adamu Shallangwa G et al (2020) Computational evaluation of potent 2 (1Himidazol- 2-yl) pyridine derivatives as potential V600E-BRAF inhibitors. Egypt J Med Hum Genet 21(1):67. https://doi.org/10.1186/s43042-020-00111-2
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7(1):42717
Benhander GM, Ashraf Ahmed Ali Abdusalam (2021) Identification of Potential Inhibitors of SARS–CoV–2 Main Protease from Allium roseum L. Molecular Docking Study. Chemistry Africa. https://doi.org/10.1007/s42250-021-00296-y
Fabian U, Shallangwa GA, Uzairu A, Abdulkadir I (2022) A combined 2D and 3D QSAR modeling, molecular docking study, design, and pharmacokinetic profiling of some arylimidamide-azole hybrids as superior L. donovani inhibitors Bul Nat Res Cent 46(189):1–24
Abdullahi SH, Uzairu A, Shallangwa GA, Sani Uba and Abdullahi Bello Umar (2022) Computational modeling, ligand–based drug design, drug–likeness and ADMET properties studies of series of chromen–2–ones analogues as anti–cancer agents. Bull Natl Res Centre 46:177
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
No potential Conflict of Interest was declared by the authors of this research work.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abdullahi, S.H., Uzairu, A., Shallangwa, G.A. et al. Structure Based Design of Some Novel 3-Methylquinoxaline Derivatives Through Molecular Docking and Pharmacokinetics Studies as Novel VEGFR-2 Inhibitors. Chemistry Africa 5, 1967–1978 (2022). https://doi.org/10.1007/s42250-022-00485-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42250-022-00485-3