Abstract
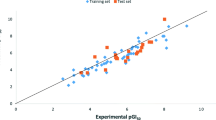
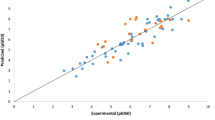
The global incidence of melanoma cancer is increasing rapidly and its metastatic form causes high mortality rates. The research report illustrates the development of a quantitative structure–activity relationship (QSAR) to predict the activities (pGI50) of some anti-cancer agents on melanoma cancer cell line. Subsequently, most potent compounds that showed better pGI50 activities were selected and screened via Lipinski’s rule of five for drug likeness, ADMET risk parameters and lastly docking simulation studies was performed to elucidate their binding mode. Kennard-Stone algorithm was adopted for the data division, the genetic function algorithm was employed for variable selection and multiple linear regressions method was utilized for model development. The derived QSAR model showed, respectively acceptable [(\(R^{2}\) (0.805), \(R_{adjusted}^{2}\) (0.773), Q2cv (0.754) and \(R_{pred}^{2}\) (0.703)]. The obtained cR2P for Y-randomization is 0.649, and applicability-domain (A-D) was assessed via leveraged method. Among the screened compounds via Lipinski’s rule of five for oral bioavailability, ADMET risk filter for drug-like features and Docking simulation studies, compound 18 and 25 were identified as the best ligands having the better interactions energies (− 150.679 kcal mol−1 and − 176.246 kcal mol−1) and showed better interactions with the receptor than vemurafenib (− 147.245 kcal mol−1). Thus, the results of this research would be helpful in identification of lead molecule and optimization of novel drug.








Similar content being viewed by others
Abbreviations
- DFT:
-
Density Functional Theory
- MVD:
-
Molegro Virtual Docker
- QSAR:
-
Quantitative structure–activity relationship
- ADMET:
-
Absorption, distribution, metabolism, excretion, and toxicity
References
Haass NK, Smalley KS, Li L, Herlyn M (2005) Adhesion, migration and communication in melanocytes and melanoma. Pigment Cell Res 18(3):150–159
Miller AJ, Mihm MC Jr (2006) Melanoma. N Engl J Med 355(1):51–65
Wolchok JD, Yuan J, Houghton AN, Gallardo HF, Rasalan TS, Wang J, Zhang Y, Ranganathan R, Chapman PB, Krown SE (2007) Safety and immunogenicity of tyrosinase DNA vaccines in patients with melanoma. Mol Ther 15(11):2044–2050
Rigon RB, Oyafuso MH, Fujimura AT, Gonçalez ML, Prado AH, Gremião MPD, Chorilli M (2015) Nanotechnology-based drug delivery systems for melanoma antitumoral therapy: a review. BioMed Res Int 2015:1–22
Ferreira LM, Cervi VF, Sari MHM, Barbieri AV, Ramos AP, Copetti PM, de Brum GF, Nascimento K, Nadal JM, Farago PV (2018) Diphenyl diselenide loaded poly (ε-caprolactone) nanocapsules with selective antimelanoma activity: development and cytotoxic evaluation. Mater Sci Eng C Mater Biol Appl 91:1
Kavanagh D, Hill A, Djikstra B, Kennelly R, McDermott E, O’Higgins N (2005) Adjuvant therapies in the treatment of stage II and III malignant melanoma. Surgeon 3(4):245–256
Grossman D, Altieri DC (2001) Drug resistance in melanoma: mechanisms, apoptosis, and new potential therapeutic targets. Cancer Metastasis Rev 20(1–2):3–11
Dubois RW, Swetter SM, Atkins M, McMasters K, Halbert R, Miller SJ, Shiell R, Kirkwood J (2001) Developing indications for the use of sentinel lymph node biopsy and adjuvant high-dose interferon alfa-2b in melanoma. Arch Dermatol 137(9):1217–1224
Kumar RS, Messina JL, Sondak EK, Reed DR (2015) Treating melanoma in adolescents and young adults: challenges and solutions. Clin Oncol Adolesc Young Adults 5:75–86
Wu C-P, Ambudkar SV (2014) The pharmacological impact of ATP-binding cassette drug transporters on vemurafenib-based therapy. Acta Pharm Sin B 4(2):105–111
Bass AS, Hombo T, Kasai C, Kinter LB, Valentin J-P (2015) A historical view and vision into the future of the field of safety pharmacology. Principles of Safety Pharmacology. Springer, New York, pp 3–45
Rodriguez B, Carusi A, Abi-Gerges N, Ariga R, Britton O, Bub G, Bueno-Orovio A, Burton RA, Carapella V, Cardone-Noott L (2015) Human-based approaches to pharmacology and cardiology: an interdisciplinary and intersectorial workshop. Ep Europace 18(9):1287–1298
Danishuddin M, Khan AU (2015) Structure based virtual screening to discover putative drug candidates: necessary considerations and successful case studies. Methods 71:135–145
Danishuddin M, Khan AU (2015) Virtual screening strategies: a state of art to combat with multiple drug resistance strains. MOJ Proteom Bioinform 2(2):00042
Shahlaei M (2013) Descriptor selection methods in quantitative structure–activity relationship studies: a review study. Chem Rev 113(10):8093–8103
Martin YC (1998) 3D QSAR: current state, scope, and limitations. 3D QSAR in drug design. Springer, Berlin, pp 3–23
Ashton PR, Fyfe MC, Hickingbottom SK, Stoddart JF, White AJ, Williams DJ (1998) Hammett correlations ‘beyond the molecule’ 1. J Chem Soc Perkin Trans 2(10):2117–2128
Verma J, Khedkar VM, Coutinho EC (2010) 3D-QSAR in drug design—a review. Curr Top Med Chem 10(1):95–115
Dhillon AS, Hagan S, Rath O, Kolch W (2007) MAP kinase signalling pathways in cancer. Oncogene 26(22):3279
Amin SA, Gayen S (2016) Modelling the cytotoxic activity of pyrazolo-triazole hybrids using descriptors calculated from the open source tool “PaDEL-descriptor”. J Taibah Univ Sci 10(6):896–905
Yap CW (2011) PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem 32(7):1466–1474
Rajer-Kanduč K, Zupan J, Majcen N (2003) Separation of data on the training and test set for modelling: a case study for modelling of five colour properties of a white pigment. Chemom Intell Lab Syst 65(2):221–229
Kennard RW, Stone LA (1969) Computer aided design of experiments. Technometrics 11(1):137–148
Tropsha A, Gramatica P, Gombar VK (2003) The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. Mol Inform 22(1):69–77
Golbraikh A, Tropsha A (2000) Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. Mol Divers 5(4):231–243
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2012) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 64:4–17
Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A (2002) BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res 62(23):6997–7000
Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G (2010) Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467(7315):596
Choi W-K, El-Gamal MI, Choi HS, Baek D, Oh C-H (2011) New diarylureas and diarylamides containing 1, 3, 4-triarylpyrazole scaffold: synthesis, antiproliferative evaluation against melanoma cell lines, ERK kinase inhibition, and molecular docking studies. Eur J Med Chem 46(12):5754–5762
Wu W, Zhang C, Lin W, Chen Q, Guo X, Qian Y, Zhang L (2015) Quantitative structure-property relationship (QSPR) modeling of drug-loaded polymeric micelles via genetic function approximation. PLoS One 10(3):e0119575
Umar BA, Uzairu A, Shallangwa GA, Sani U (2019) QSAR modeling for the prediction of pGI50 activity of compounds on LOX IMVI cell line and ligand-based design of potent compounds using in silico virtual screening. Netw Model Anal Health Inform Bioinform 8(1):22
Gadaleta D, Mangiatordi GF, Catto M, Carotti A, Nicolotti O (2016) Applicability domain for QSAR models: where theory meets reality. Int J Quant Struct Prop Relatsh (IJQSPR) 1(1):45–63
Roy K, Kar S, Das RN (2015) A primer on QSAR/QSPR modeling: fundamental concepts. Springer, New York
Gramatica P, Giani E, Papa E (2007) Statistical external validation and consensus modeling: a QSPR case study for Koc prediction. J Mol Graph Model 25(6):755–766
Jalali-Heravi M, Konuze E (2002) Use of quantitative structure property relationships in predicting the Kraft point of anionic surfactants. Electron J Mol Des 1:410–417
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717
Montanari F, Ecker GF (2015) Prediction of drug–ABC-transporter interaction—recent advances and future challenges. Adv Drug Deliv Rev 86:17–26
Szakacs G, Varadi A, Özvegy-Laczka C, Sarkadi B (2008) The role of ABC transporters in drug absorption, distribution, metabolism, excretion and toxicity (ADME–Tox). Drug Discov Today 13(9–10):379–393
Guimarães CR, Rai BK, Munchhof MJ, Liu S, Wang J, Bhattacharya SK, Buckbinder L (2011) Understanding the impact of the P-loop conformation on kinase selectivity. J Chem Inf Model 51(6):1199–1204
Adedirin O, Uzairu A, Shallangwa GA, Abechi SE (2018) Optimization of the anticonvulsant activity of 2-acetamido-N-benzyl-2-(5-methylfuran-2-yl) acetamide using QSAR modeling and molecular docking techniques. Beni-Suef Univ J Basic Appl Sci 7(4):430–440
Umar BA, Uzairu A, Shallangwa GA, Uba S (2019) Rational drug design of potent V600E-BRAF kinase inhibitors through molecular docking simulation. J Eng Exact Sci 5(5):0469–0481
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors report no conflict of interest.
Rights and permissions
About this article
Cite this article
Umar, A.B., Uzairu, A., Uba, S. et al. QSAR and Docking Studies on Some Potential Anti-Cancer Agents to Predict their Effect on M14 Melanoma Cell Line. Chemistry Africa 3, 1009–1022 (2020). https://doi.org/10.1007/s42250-020-00185-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42250-020-00185-w