Abstract
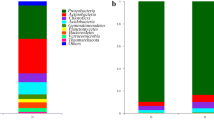
Tobacco mosaic virus (TMV) is one of the most economically damaging plant viruses because of its wide host range and transmission mode. TMV manipulates the plant to produce viral proteins, instead of resistant ones. However, microbes with pathogenic defenses have been beneficial against the biotic stress caused by TMV. Through DNA metabarcoding, the microbe communities between parts of the phyllosphere and rhizosphere infected by TMV were compared over a two-week period. Overall, there was a variation of microbial presence and higher species richness in the mock-inoculated than viral inoculated plants. After one week, the mock inoculated plant organs had higher species richness than viral inoculated plant organs due to the presence of viral threat. After two weeks, the viral inoculated soil had the second highest species richness due to the soil being colonized and the presence of defense mechanisms. In addition, there was a wide range of taxa present such as Proteobacteria, Bacteroidetes, Acidobacteria, and Cyanobacteria.





Similar content being viewed by others
Abbreviations
- TMV:
-
Tobacco Mosaic Virus
- UV:
-
Ultraviolet
- C:
-
Control/Mock Inoculated
- T:
-
Experimental/Viral Inoculated
- L1:
-
Inoculated Leaf 1
- L2:
-
Inoculated Leaf 2
- ST:
-
Systemic Leaf
- S:
-
Soil
- R:
-
Root
- GFP:
-
Green Fluorescent Protein
References
Bagley CA (2001) Controlling tobacco mosaic virus in tobacco through resistance. Retrieved May 21, 2019. from https://vtechworks.lib.vt.edu/handle/10919/30911
Boller T, He SY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science (New York, N.Y.) 324(5928):742–744. https://doi.org/10.1126/science.1171647
Brewer HC, Hird DL, Bailey AM, Seal SE, Foster GD (2017). A guide to the contained use of plant virus infectious clones. Plant Biotechnol J 832–843. https://doi.org/10.1111/pbi.12876
Catão EC, Lopes FA, Araújo JF, de Castro AP, Barreto CC, Bustamante MM, Quirino BF, Krüger RH (2014) Soil acidobacterial 16S rrna gene sequences reveal subgroup level differences between savanna-like Cerrado and Atlantic Forest Brazilian biomes. Int J Microbiol 201(4):1–12. https://doi.org/10.1155/2014/156341
Chauhan P, Singla K, Rajbhar M, Singh A, Das N, Kumar K (2019) A systematic review of conventional and advanced approaches for the control of plant viruses. J Appl Biol Biotechnol 7:89–98. https://doi.org/10.7324/JABB.2019.70414
Creager AN (1999) Tobacco mosaic virus: pioneering research for a century. The Plant Cell Online 11(3):301–308. https://doi.org/10.1105/tpc.11.3.301
Enebe MC, Babalola OO (2018) The impact of microbes in the orchestration of plants’ resistance to biotic stress: a disease management approach. Appl Microbiol Biotechnol 103(1):9–25. https://doi.org/10.1007/s00253-018-9433-3
Franco-Frías E, Mercado-Guajardo V, Merino-Mascorro A, Pérez-Garza J, Heredia N, León JS, Jaykus L-A, Dávila-Aviña J, García S (2021) Analysis of bacterial communities by 16S rrna gene sequencing in a melon-producing agro-environment. Microb Ecol 82(3):613–622. https://doi.org/10.1007/s00248-021-01709-8
Fry JC, Webster G, Cragg BA, Weightman AJ, Parkes RJ (2006) Analysis of DGGE profiles to explore the relationship between prokaryotic community composition and biogeochemical processes in deep subseafloor sediments from the peru margin. FEMS Microbiol Ecol 58(1):86–98. https://doi.org/10.1111/j.1574-6941.2006.00144.x
Fujii T, Minami M, Watanabe T, Sato T, Kumaishi K, Ichihashi Y (2021) Characterization of inter-annual changes in soil microbial flora of Panax ginseng cultivation fields in Shimane Prefecture of western Japan by DNA metabarcoding using next-generation sequencing. J Nat Med 75(4):1067–1079. https://doi.org/10.1007/s11418-021-01514-0
Goodsell D (2003) PDB101: Molecule of the month: Green Fluorescent Protein (GFP). Retrieved May 5, 2019 from https://pdb101.rcsb.org/motm/42
Goodsell D (2009) PDB101: Molecule of the month: tobacco mosaic virus. Retrieved May 5, 2019 from https://pdb101.rcsb.org/motm/109
Gower JC (1966) Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53(3/4):325. https://doi.org/10.2307/2333639
Hossain D (2020) Determination of economic Threshold Level (ETL) of tobacco mosaic and leaf curl viruses. Int J Sci: Basic and Appl Res (IJSBAR) 43:147–165
Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ (2001) Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol 67(10):4399–4406. https://doi.org/10.1128/AEM.67.10.4399-4406.2001
Hull R (2009) Mechanical inoculation of plant viruses. Curr Prot Microbiol. Retrieved May 5, 2019 from https://pubmed.ncbi.nlm.nih.gov/19412912/
Kielak AM, Cipriano MA, Kuramae EE (2016) Acidobacteria strains from subdivision 1 act as Plant Growth-promoting bacteria. Arch Microbiol 198(10):987–993. https://doi.org/10.1007/s00203-016-1260-2
Lamichhane JR, Venturi V (2015) Synergisms between microbial pathogens in plant disease complexes: A growing trend. Front Plant Sci 6. https://doi.org/10.3389/fpls.2015.00385
Lidbury ID, Borsetto C, Murphy AR, Bottrill A, Jones AM, Bending GD, Hammond JP, Chen Y, Wellington EM, Scanlan DJ (2020) Niche-adaptation in plant-associated bacteroidetes favours specialisation in organic phosphorus mineralisation. ISME J 15(4):1040–1055. https://doi.org/10.1038/s41396-020-00829-2
Nadkarni MA, Hunter N, Jacques NA, Martin FE (2002) Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148(1):257–266. https://doi.org/10.1099/00221287-148-1-257
Nannipieri P, Ascher J, Ceccherini MT, Landi L, Pietramellara G, Renella G (2003) Microbial diversity and soil functions. Eur J Soil Sci 54(4):655–670. https://doi.org/10.1046/j.1351-0754.2003.0556.x
Nosek BA, Errington TM (2020) What is replication?. PLoS Biol 18(3):e3000691
Ogaki MB, Pinto OH, Vieira R, Neto AA, Convey P, Carvalho-Silva M, Rosa CA, Câmara PE, Rosa LH (2021) Fungi present in Antarctic deep-sea sediments assessed using DNA metabarcoding. Microb Ecol 82(1):157–164. https://doi.org/10.1007/s00248-020-01658-8
Peng J, Song K, Zhu H, Kong W, Liu F, Shen T, He Y (2017) Fast detection of tobacco mosaic virus infected tobacco using laser-induced breakdown spectroscopy. Sci Rep 7(1). https://doi.org/10.1038/srep44551
Peng S, Schmid B, Haase J, Niklaus PA (2016) Leaf area increases with species richness in young experimental stands of subtropical tree s. J Plant Ecol 10(1):128–135. https://doi.org/10.1093/jpe/rtw016
Rai AN, Singh AK, Syiem MB (2019) Plant growth-promoting abilities in cyanobacteria. Cyanobacteria 459–476. https://doi.org/10.1016/b978-0-12-814667-5.00023-4
Rao IM, Miles JW, Beebe SE, Horst WJ (2016) Root adaptations to soils with low fertility and aluminium toxicity. Ann Bot 118(4):593- 605. https://doi.org/10.1093/aob/mcw073
Sousa LP, Silva MJ, Costa Mondego JM (2018) Leaf-associated bacterial microbiota of coffee and its correlation with manganese and calcium levels on leaves. Genet Mol Biol 41(2):455–465. https://doi.org/10.1590/1678-4685-gmb-2017-0255
Tawfik SA, Azab MM, Ahmed AA, Fayyad DM (2018) Illumina MiSeq sequencing for preliminary analysis of microbiome causing primary endodontic infections in Egypt. Int J Microbiol 2018:1–15. https://doi.org/10.1155/2018/2837328
Wang F, Men X, Zhang G, Liang K, Xin Y, Wang J, Wu L (2018) Assessment of 16S rRNA gene primers for studying bacterial communit y structure and function of aging flue-cured tobaccos. AMB Express 8(1). https://doi.org/10.1186/s13568-018-0713-1
Wei Z, Hu X, Li X, Zhang Y, Jiang L, Li J, Guan Z, Cai Y, Liao X (2017) The rhizospheric microbial community structure and diversity of deciduous and evergreen forests in Taihu Lake area, China. PloS One 12(4). https://doi.org/10.1371/journal.pone.0174411
Acknowledgements
The authors would like to thank the amazing contributors who have made this research possible, starting from the staff at Cold Spring Harbor Laboratory DNA Learning Center and Stony Brook University. The researchers who made this research successful are Dr. Sharon Pepenella, Dr. Cristina Fernandez-Marco, Dr. Joslynn Lee, Dr. Cornel Ghiban, Dr. David Micklos, and Dr. Benoit Lacroix. Within the William Floyd High School’s Administration and Research Program, Ms. Christine Rosado, Mr. Philip Scotto, Ms. Lisa Pisano, Mrs. Deborah Gurney, Ms. Kathleen Keane, and Mr. Kevin Coster have provided an abundant amount of support and resources which have influenced this research in a positive way.
Funding
Research was supported by the DNA Learning Center at Cold Spring Harbor Laboratory. Bruce Nash is associated with this laboratory.
Author information
Authors and Affiliations
Contributions
This research study was designed and conducted by C.H.,V.H., and P.T. B.N., and P.T. managed and coordinated responsibility for the research activity and execution. V.H., L.M., and P.T. provided mentorship and oversight. B.N. provided the funding and reagents. P.T. provided the genetically engineered TMV and his glass chambers to grow the Nicotiana benthamiana plants in. The manuscript was written by C.H., and revised by V.H., L.M., P.T., and B.N.
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hossain, C., Hernandez, V., McHugh, L. et al. Analyzing the impacts of tobacco mosaic virus on the microbial diversity of Nicotiana benthamiana. J Plant Pathol 104, 959–967 (2022). https://doi.org/10.1007/s42161-022-01103-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42161-022-01103-4