Abstract
Purpose
To develop and validate an artificial intelligence (AI)-assisted framework for fast and automatic radiofrequency ablation (RFA) planning of liver tumors from CT images.
Materials and methods
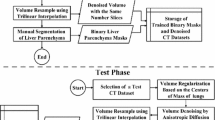
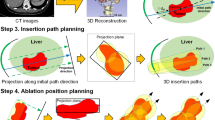
This framework consisted of three steps. First, the abdominal multi-organs related to RFA planning were automatically segmented from CT images using a modified nnU-Net with class-weighted loss function. Then, utilizing the segmented liver as a location prior, the liver tumors and hepatic vessels were further segmented by a sensitivity-enhanced segmentation network. Finally, a clinically acceptable RFA plan was generated by a fully automatic planning method based on the segmented organs and tumors. Experiments were conducted on two public segmentation datasets and patients from two different centers to evaluate the proposed framework.
Results
The proposed abdominal multi-organ segmentation model achieved an average dice of 87.7 \(\pm\) 8.0% on 15 abdominal organs and the proposed liver tumor and hepatic vessel segmentation model achieved an average dice of 80.7 \(\pm\) 11.2% and 65.3 \(\pm\) 10.2% and an average sensitivity of 87.4 \(\pm\) 12.9% and 76.8 \(\pm\) 14.9% for liver tumor and hepatic vessel, respectively. Finally, the proposed framework generated clinically acceptable RFA plans within a few minutes without human intervention for all patients from two centers.
Conclusion
The proposed AI-assisted framework is fast and can automatically generate clinically acceptable RFA plans for liver tumors from CT images, which can assist interventional radiologists in determining suitable plans and reduce their burden.




Similar content being viewed by others
Data availability
Data generated or analyzed during the study are not openly available due to reasons of sensitivity and are available from the corresponding author upon reasonable request.
References
Sun L, Yang Y, Li Y, Zhang B, Shi R. The past, present, and future of liver cancer research in China. Cancer Lett. 2023;2023: 216334. https://doi.org/10.1016/j.canlet.2023.216334.
Garrean S, Hering J, Saied A, Helton WS, Espat NJ. Radiofrequency ablation of primary and metastatic liver tumors: a critical review of the literature. Am J Surg. 2008;195:508–20. https://doi.org/10.1016/j.amjsurg.2007.06.024.
Benson AB 3rd, Abrams TA, Ben-Josef E, et al. NCCN clinical practice guidelines in oncology: hepatobiliary cancers. J Natl Compr Cancer Netw: JNCCN. 2009;7:350–91. https://doi.org/10.6004/jnccn.2009.0027.
Chen MH, Yang W, Yan K, et al. Large liver tumors: protocol for radiofrequency ablation and its clinical application in 110 patients—mathematic model, overlapping mode, and electrode placement process. Radiology. 2004;232:260–71. https://doi.org/10.1148/radiol.2321030821.
Schumann C, Rieder C, Bieberstein J, et al. State of the art in computer-assisted planning, intervention, and assessment of liver tumor ablation. Crit Rev™ Biomed Eng. 2010;38:31–52. https://doi.org/10.1615/CritRevBiomedEng.v38.i1.40.
Minami Y, Kudo M. Radiofrequency ablation of hepatocellular carcinoma: a literature review. Int J Hepatol. 2011;2011:1–9. https://doi.org/10.4061/2011/104685.
Wang Y, Zhou Y, Shen W, Park S, Fishman EK, Yuille AL. Abdominal multi-organ segmentation with organ-attention networks and statistical fusion. Med Image Anal. 2019;55:88–102. https://doi.org/10.1016/j.media.2019.04.005.
Hu P, Wu F, Peng J, Bao Y, Chen F, Kong D. Automatic abdominal multi-organ segmentation using deep convolutional neural network and time-implicit level sets. Int J Comput Ass Rad. 2017;12:399–411. https://doi.org/10.1007/s11548-016-1501-5.
Chen H, Wang X, Huang Y, Wu X, Yu Y, Wang L (2019) Harnessing 2D networks and 3D features for automated pancreas segmentation from volumetric CT images. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019, pp 339–347. https://doi.org/10.1007/978-3-030-32226-7_38
Gul S, Khan MS, Bibi A, Khandakar A, Ayari MA, Chowdhury ME. Deep learning techniques for liver and liver tumor segmentation: a review. Comput Biol Med. 2022;147: 105620. https://doi.org/10.1016/j.compbiomed.2022.105620.
Huang YJ, Dou Q, Wang ZX, et al. 3-D RoI-aware U-net for accurate and efficient colorectal tumor segmentation. IEEE T Cybern. 2020;51(11):5397–408. https://doi.org/10.1109/TCYB.2020.2980145.
Li R, Huang YJ, Chen H, et al. 3d graph-connectivity constrained network for hepatic vessel segmentation. IEEE J Biomed Health. 2021;26(3):1251–62. https://doi.org/10.1109/JBHI.2021.3118104.
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, pp 234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Isensee F, Jaeger PF, Kohl SA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18:203–11. https://doi.org/10.1038/s41592-020-01008-z.
Chaitanya K, Audigier C, Balascuta LE, Mansi T (2022) Automatic planning of liver tumor thermal ablation using deep reinforcement learning. In: International conference on medical imaging with deep learning, PMLR 172, pp 219–230
Meister F, Audigier C, Passerini T, et al. Fast automatic liver tumor radiofrequency ablation planning via learned physics model. In: Medical image computing and computer assisted intervention. Cham: Springer; 2022. p. 167–76. https://doi.org/10.1007/978-3-031-16449-1_17.
Liang L, Cool D, Kakani N, Wang G, Ding H, Fenster A. Automatic radiofrequency ablation planning for liver tumors with multiple constraints based on set covering. IEEE Trans Med Imaging. 2019;39:1459–71. https://doi.org/10.1109/TMI.2019.2950947.
Yu P, Fu T, Wu C, Jiang Y, Yang J (2021) Automatic radiofrequency ablation planning for liver tumors: a planning method based on the genetic algorithm with multiple constraints. In: 2021 the 3rd International conference on intelligent medicine and health. Association for Computing Machinery, pp 8–14. https://doi.org/10.1145/3484377.3484379
Li R, An C, Wang S, et al. A heuristic method for rapid and automatic radiofrequency ablation planning of liver tumors. Int J Comput Ass Rad. 2023;2023:1–9. https://doi.org/10.1007/s11548-023-02921-2.
Seror O. Ablative therapies: advantages and disadvantages of radiofrequency, cryotherapy, microwave and electroporation methods, or how to choose the right method for an individual patient? Diagn Interv Imaging. 2015;96:617–24. https://doi.org/10.1016/j.diii.2015.04.007.
Ji Y, Bai H, Ge C, et al. Amos: A large-scale abdominal multi-organ benchmark for versatile medical image segmentation. Adv Neural Inf Process Syst. 2022;35:36722–32.
Antonelli M, Reinke A, Bakas S, et al. The medical segmentation decathlon. Nat Commun. 2022;13(1):4128. https://doi.org/10.1038/s41467-022-30695-9.
Laimer G, Schullian P, Jaschke N, et al. Minimal ablative margin (MAM) assessment with image fusion: an independent predictor for local tumor progression in hepatocellular carcinoma after stereotactic radiofrequency ablation. Eur Radiol. 2020;30:2463–72. https://doi.org/10.1007/s00330-019-06609-7.
Funding
This study was supported by grants from the National Key Research and Development Program of China (2019YFC0118100), National Natural Science Foundation of China (62171167).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This retrospective study was conducted following ethical approval from the Institutional Review Board of The Third Medical Center of Chinese PLA General Hospital and Daqing Longnan Hospital, and all CT images are anonymized. This paper does not contain any studies with animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, R., Xin, R., Wang, S. et al. An artificial intelligence-assisted framework for fast and automatic radiofrequency ablation planning of liver tumors in CT images. Chin J Acad Radiol (2024). https://doi.org/10.1007/s42058-024-00145-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42058-024-00145-0