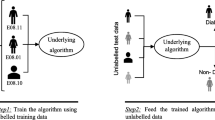
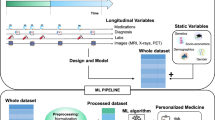
Abstract
Longitudinal disease subtyping is an important problem within the broader scope of computational phenotyping. In this article, we discuss several data-driven unsupervised disease subtyping methods to obtain disease subtypes from longitudinal clinical data. The methods are analyzed in the context of chronic kidney disease, one of the leading health problems, both in the USA and worldwide. To provide a quantitative comparison of the different methods, we propose a novel evaluation metric that measures the cluster tightness and degree of separation between the various clusters produced by each method. Comparative results for two significantly large clinical datasets are provided, along with key insights that are possible due to the proposed evaluation metric.







Similar content being viewed by others
References
Baytas IM, Xiao C, Zhang X, Wang F, Jain AK, Zhou J (2017) Patient subtyping via time-aware LSTM networks. In: Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining. ACM, pp 65–74
Choi E, Bahadori MT, Searles E, Coffey C, Thompson M, Bost J, Tejedor-Sojo J, Sun J (2016) Multi-layer representation learning for medical concepts. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. ACM, pp 1495–1504
Choi E, Xiao C, Stewart W, Sun J (2018) Mime: multilevel medical embedding of electronic health records for predictive healthcare. In: Advances in neural information processing systems, pp 4547–4557
Hripcsak G, Albers DJ (2012) Next-generation phenotyping of electronic health records. J Am Med Inform Assoc 20(1):117–121
Jain AK, Murty MN, Flynn PJ (1999) Data clustering: a review. ACM Comput Surv (CSUR) 31(3):264–323
Johnson AE, Pollard TJ, Shen L, Lehman L-wH, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG (2016) MIMIC-III, a freely accessible critical care database. Sci Data, 3
Lasko TA, Denny JC, Levy MA (2013) Correction: computational phenotype discovery using unsupervised feature learning over noisy, sparse, and irregular clinical data. PLoS one 8:8
Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T et al (2009) A new equation to estimate glomerular filtration rate. Ann Intern Med 150(9):604–612
Liao TW (2005) Clustering of time series data - a survey. Pattern Recogn 38 (11):1857–1874
Liu Y, Li Z, Xiong H, Gao X, Wu J (2010) Understanding of internal clustering validation measures. In: 2010 IEEE international conference on data mining. IEEE, pp 911–916
Luong DTA, Chandola V (2017) A k-means approach to clustering disease progressions. In: 2017 IEEE International conference on healthcare informatics (ICHI), pp 268–274
Luong DTA, Chandola V (2019) Learning deep representations from clinical data for chronic kidney disease. In: 2019 IEEE International conference on healthcare informatics (ICHI)
Luong DTA, Tran D, Pace WD, Dickinson M, Vassalotti J, Carroll J, Withiam-Leitch M, Yang M, Satchidanand N, Staton E et al (2017) Extracting deep phenotypes for chronic kidney disease using electronic health records. eGEMs (Generating Evidence & Methods to improve patient outcomes), 5
National Kidney Foundation (2002) K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification, vol 39
Pace WD, Fox C, White T, Graham D, Schilling LM, David R (2014) The DARTNet institute: seeking a sustainable support mechanism for electronic data enabled research networks. eGEMs (Generating Evidence & Methods to improve patient outcomes) 2(2):6
Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, Liu PJ, Liu X, Marcus J, Sun M et al (2018) Scalable and accurate deep learning with electronic health records. NPJ Digit Med 1(1):18
Rousseeuw PJ (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20:53–65
Schulam P, Arora R (2016) Disease trajectory maps. In: Advances in neural information processing systems, pp 4709–4717
Schulam P, Saria S (2015) A framework for individualizing predictions of disease trajectories by exploiting multi-resolution structure. In: Advances in neural information processing systems, pp 748–756
Schulam P, Wigley F, Saria S (2015) Clustering longitudinal clinical marker trajectories from electronic health data: applications to phenotyping and endotype discovery. In: AAAI, pp 2956–2964
Singh P, Chandola V, Fox C (2017) Automatic extraction of deep phenotypes for precision medicine in chronic kidney disease. In: Proceedings of the 2017 international conference on digital health. ACM, pp 195–199
Zaki MJ, Meira W Jr (2014) Data mining and analysis: fundamental concepts and algorithms. Cambridge University Press
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Luong, D.T.A., Singh, P., Ramezani, M. et al. longSil: an Evaluation Metric to Assess Quality of Clustering Longitudinal Clinical Data. J Healthc Inform Res 3, 441–459 (2019). https://doi.org/10.1007/s41666-019-00058-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41666-019-00058-z