Abstract
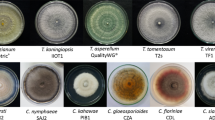
Antagonistic potentials of Trichoderma isolates were evaluated against the phytopathogen Sclerotium rolfsii causing stem rot in groundnut. The in vitro growth inhibition of test pathogen on potato dextrose agar medium was observed maximum by antagonist T. virens NBAII Tvs 12 followed by T. koningii MTCC 796 at 6 days after inoculation (DAI). The most effective antagonists T. virens NBAII Tvs 12 and T. koningii MTCC 796 exhibited different patterns of growth inhibition of test pathogen. The Tvs 12 isolate overgrew S. rolfsii by degrading mycelia through mycoparasitism during 6–12 DAI. Isolate MTCC 796 inhibited the growth of the pathogen by forming an inhibition zone (antibiosis). The biogenesis (formation) of sclerotia of S. rolfsii was elevated during weak antagonism and diminished in interactions with strong antagonists. Trichoderma antagonists were examined for molecular fingerprints and diversity using 23 ISSR primers which amplified 296 amplicons. Among them, 23 showed a unique polymorphism. Jaccard’s similarity coefficient ranged from 0.14 to 0.71, and clustering pattern exhibited minimum 22 % similarity between the mycoparasitic best antagonist Tvs12 and other isolates with antagonistic activity within Bi(a) cluster. The molecular fingerprints UBC-808(1221) and UBC-827(2123) were specific for Tvs 12 strain; and ISSR-5(1956) and UBC-812(1689) were specific for MTTC 796.



Similar content being viewed by others
References
Anderson, J. A., Churchill, G. A., Sutrique, J. E., Tanksley, S. D., & Sorrels, M. E. (1993). Optimizing parental selection for genetic linkage maps. Genome, 36, 181–186.
Catalano, V., Vergara, M., Hauzenberger, J. R., Seiboth, B., Sarrocco, S., & Vannacci, G. (2011). Use of a non-homologous end-joining-deficient strain (delta-ku70) of the biocontrol fungus Trichoderma virens to investigate the function of the laccase gene lcc1 in sclerotia degradation. Current Genetics, 57, 13–23.
Chaverri, P., Branco-Rocha, F., Jaklitsch, W., Gazis, R., Degenkolb, T., & Samuels, G. J. (2015). Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia, 107(3), 558–590.
Chet, I. (1987). Trichoderma—Application, mode of action, and potential as biocontrol agent of soilborne plant pathogenic fungi. In I. Chet (Ed.), Innovative approaches to plant disease control (pp. 37–160). New York: Wiley.
Dennis, C. J., & Webster, J. (1971). Antagonism properties of species groups of Trichoderma, III. Hyphal interaction. Transactions British Mycological Society, 57, 363–369.
Fisher, R. A., & Yates, F. (1948). Statistical tables for biological, agricultural and medical research (3rd ed., pp. 26–27). Edinburgh: Oliver and Boyd.
Gajera, H. P., Bambharolia, R. M., Patel, S. V., Mandavia, M. K., & Golakiya, B. A. (2009). Significance of lytic enzymes from Trichoderma in the in vitro biocontrol of fungal plant pathogen Fusarium oxysporum f. sp. ciceri. Indian Journal of Agricultural Biochemistry, 22(1), 31–37.
Ganesan, S., & Sekar, R. (2004). Biocontrol mechanism of groundnut (Arachis hypogaea L.) diseases-Trichoderma system. Biotechnological Applications in Environment and Agriculture, 72, 312–327.
Georgiou, C. D., Patsoukis, Ν., Papapostolou, Ι., & Zervoudakis, G. (2006). Sclerotial metamorphosis in filamentous fungi is induced by oxidative stress. Integrative and Comparative Biology, 46, 691–712.
Ghislain, M., Zhang, D., Fajardo, D., Huamann, Z., & Hijmans, R. H. (1999). Marker assisted sampling of the cultivated Andean potato Solanum phureja collection using RAPD markers. Genetic Resources and Crop Evolution, 46, 547–555.
Hirpara, D. G., Gajera, H. P., Bhimani, R. D., & Golakiya, B. A. (2016). The SRAP based molecular diversity related to antifungal and antioxidant bioactive constituents for biocontrol potentials of Trichoderma against Sclerotium rolfsii Sacc. Current Genetics, 62, 619–641.
Imtiaj, A., & Lee, T. S. (2008). Antagonistic effect of three Trichoderma species on the Alternaria porri pathogen of onion blotch. World Journal on Agricultural Sciences, 4(1), 13–17.
Joshi, D., Hooda, K. S., & Bhatt, J. C. (2008). Biocontrol potential of fungal antagonists from western Himalayan region against sclerotial plant pathogens. Pesticide Research Journal, 20(1), 1–5.
Kannan, N., Rubeena, M., Sajith, S., Sreedevi, S., Priji, P., & Unni, N. (2012). A novel strain of Trichoderma viride shows complete lignocellulolytic activities. Advances in Bioscience and Biotechnology, 3, 1160–1166.
Kumar, M. A., & Sharma, P. (2011). Molecular and morphological characters: An appurtenance for antagonism in Trichoderma spp. African Journla of Biotechnology, 22, 4532–4543.
Lewis, J. A., & Papavizas, G. C. (1987). Application of Trichoderma and Gliocladium in alginate pellets for control of Rhizoctonia damping-off. Plant Pathology, 36, 438–446.
Mukherjee, P. K. (1999). Trichoderma species as microbial suppressive agents of plant pathogens. In M. N. Jha (Ed.), Current trends in life sciences—Agromicrobes (vol. 23, pp. 261–280). New Delhi: Today and Tomorrow’s Publishers.
Narayanasamy, P. D., & Saravana, M. (2009). RAPD analysis of Trichoderma and it’s antagonistic affect with Pseudomonas fluorescens. Journal of Plant Pathology & Microbiology, 20(4), 223–233.
Naseby, D. C., Pascual, J. A., & Lynch, J. M. (1999). Effect of biocontrol strains of Trichoderma on plant growth, Pythium ultimum populations, soil microbial communities and soil enzyme activities. Journal of Applied Microbiology, 88, 161–169.
Nei, M. (1987). Molecular evolutionary genetics. New York: Columbia University Press.
Pan, S., & Bhagat, S. (2007). Effect of substrates physical factors on mass multiplication of Trichoderma harzianum in management of seedling blight of jute. Biological Control, 21, 127–136.
Papavizas, C. G. (1985). Trichoderma and Gliocladium: Biology, ecology, and potential for biocontrol. Annual Review of Phytopathology, 23, 23–54.
Pérez, G., Verdejo, V., Gondim-porto, C., Orlando, J., & Carú, M. (2014). Designing a SCAR molecular marker for monitoring Trichoderma cf. harzianum in experimental communities. Journal of Zhejiang University Sciences B (Biomedicine & Biotechnology), 15(11), 966–978.
Rohlf, F. J. (1998). Numerical taxonomy and multivariate analysis system version 1.80 manual. New York: Exeter Software.
Samuels, G. J. (1996). Trichoderma: A review of biology and systematics of the genus. Mycological Research, 21, 420–432.
Samuels, G. J., Pardo-Schultheiss, R., Hebbar, P., Lumsden, R. D., Bastos, C. N., & Costa, J. C. (2000). Trichoderm stromaticum, sp. nov., a parasite of the cacao witches broom pathogen. Mycological Research, 104, 760–764.
Sarroccoa, S., Mikkelsenb, L., Vergarac, M., Jensenb, D., Beckb, M., & Vannaccia, G. (2006). Histopathological studies of sclerotia of phytopathogenic fungi parasitized by a GFP transformed Trichoderma virens antagonistic strain. Mycological Research, 110, 179–187.
Sinclair, J. B., & Dhingra, O. D. (1985). Basic plant pathology methods. Boca Rotam, Florida: CRC Press, Inc.
Sivan, A., & Chet, I. (1989). Degradation of fungal cell walls by lytic enzymes of Trichoderma harzianum. Journal of General Microbiology, 135, 675–682.
Sobral, B. W., & Honeycutt, R. J. (1993). High output genetic mapping of polyploids using PCR-generated markers. Theoretical and Applied Genetics, 86, 105–112.
Yang, X., & Quiros, C. F. (1995). Construction of a genetic linkage map in celery using DNA-based markers. Genome, 38, 36–44.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hirpara, D.G., Gajera, H.P., Hirpara, H.Z. et al. Molecular diversity and fingerprints of Trichoderma associated with antagonistic potentials against Sclerotium rolfsii Sacc.. J Plant Dis Prot 124, 31–40 (2017). https://doi.org/10.1007/s41348-016-0053-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41348-016-0053-9