Abstract
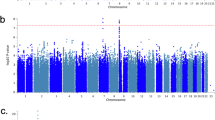
Idiopathic hypersomnia (IH) is a rare sleep disorder characterized by excessive daytime sleepiness, great difficulty upon awakening, and prolonged sleep time. In contrast to narcolepsy type 1, which is a well-recognized hypersomnia, the etiology of IH remains poorly understood. No susceptibility loci for IH have been identified, although familial aggregations have been observed among patients with IH. Narcolepsy type 1 is strongly associated with human leukocyte antigen (HLA)-DQB1*06:02; however, no significant associations between IH and HLA alleles have been reported. To identify genetic variants that affect susceptibility to IH, we performed a genome-wide association study (GWAS) and two replication studies involving a total of 414 Japanese patients with IH and 6587 healthy Japanese individuals. A meta-analysis of the three studies found no single-nucleotide polymorphisms (SNPs) that reached the genome-wide significance level. However, we identified several candidate SNPs for IH. For instance, a common genetic variant (rs2250870) within an intron of PDE9A was suggestively associated with IH. rs2250870 was significantly associated with expression levels of PDE9A in not only whole blood but also brain tissues. The leading SNP in the PDE9A region was the same in associations with both IH and PDE9A expression. PDE9A is a potential target in the treatment of several brain diseases, such as depression, schizophrenia, and Alzheimer’s disease. It will be necessary to examine whether PDE9A inhibitors that have demonstrated effects on neurophysiologic and cognitive function can contribute to the development of new treatments for IH, as higher expression levels of PDE9A were observed with regard to the risk allele of rs2250870. The present study constitutes the first GWAS of genetic variants associated with IH. A larger replication study will be required to confirm these associations.



Similar content being viewed by others
References
Billiard M, Sonka K. Idiopathic hypersomnia. Sleep Med Rev. 2016;29:23–33.
Ali M, Auger RR, Slocumb NL, Morgenthaler TI. Idiopathic hypersomnia: clinical features and response to treatment. J Clin Sleep Med. 2009;5:562–8.
Anderson KN, Pilsworth S, Sharples LD, et al. Idiopathic hypersomnia: a study of 77 cases. Sleep. 2007;30:1274–81.
Bassetti C, Aldrich MS. Idiopathic hypersomnia. A series of 42 patients. Brain. 1997;120(Pt 8):1423–35.
Arnulf I, Leu-Semenescu S, Dodet P. Precision medicine for idiopathic hypersomnia. Sleep Med Clin. 2019;14:333–50.
Trotti LM. Idiopathic hypersomnia. Sleep Med Clin. 2017;12:331–44.
Juji T, Satake M, Honda Y, Doi Y. HLA antigens in Japanese patients with narcolepsy. All the patients were DR2 positive. Tissue Antigens. 1984;24:316–9.
Mignot E, Lin L, Rogers W, et al. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am J Hum Genet. 2001;68:686–99.
Miyagawa T, Toyoda H, Kanbayashi T, et al. An association analysis of HLA-DQB1 with narcolepsy without cataplexy and idiopathic hypersomnia with/without long sleep time in a Japanese population. Hum Genome Var. 2015;2:15031.
Nishino S, Ripley B, Overeem S, et al. Hypocretin (orexin) deficiency in human narcolepsy. Lancet. 2000;355:39–40.
Mignot E, Lammers GJ, Ripley B, et al. The role of cerebrospinal fluid hypocretin measurement in the diagnosis of narcolepsy and other hypersomnias. Arch Neurol. 2002;59:1553–62.
Tam V, Patel N, Turcotte M, et al. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–84.
Miyagawa T, Kawashima M, Nishida N, et al. Variant between CPT1B and CHKB associated with susceptibility to narcolepsy. Nat Genet. 2008;40:1324–8.
Hallmayer J, Faraco J, Lin L, et al. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat Genet. 2009;41:708–11.
Miyagawa T, Khor SS, Toyoda H, et al. A variant at 9q34.11 is associated with HLA-DQB1*06:02 negative essential hypersomnia. J Hum Genet. 2018;63:1259–67.
Tadaka S, Katsuoka F, Ueki M, et al. 3.5KJPNv2: an allele frequency panel of 3552 Japanese individuals including the X chromosome. Hum Genome Var. 2019;6:28.
Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82.
Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9.
Miyagawa T, Nishida N, Ohashi J, et al. Appropriate data cleaning methods for genome-wide association study. J Hum Genet. 2008;53:886–93.
Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5: e1000529.
Marchini J, Howie B, Myers S, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13.
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5.
Nishida N, Tanabe T, Takasu M, et al. Further development of multiplex single nucleotide polymorphism typing method, the DigiTag2 assay. Anal Biochem. 2007;364:78–85.
Mele M, Ferreira PG, Reverter F, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–5.
Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7.
Fromer M, Roussos P, Sieberts SK, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–53.
Fisher DA, Smith JF, Pillar JS, et al. Isolation and characterization of PDE9A, a novel human cGMP-specific phosphodiesterase. J Biol Chem. 1998;273:15559–64.
Kleiman RJ, Chapin DS, Christoffersen C, et al. Phosphodiesterase 9A regulates central cGMP and modulates responses to cholinergic and monoaminergic perturbation in vivo. J Pharmacol Exp Ther. 2012;341:396–409.
Yoshida K, Tsunoda SP, Brown LS, Kandori H. A unique choanoflagellate enzyme rhodopsin exhibits light-dependent cyclic nucleotide phosphodiesterase activity. J Biol Chem. 2017;292:7531–41.
Langmesser S, Franken P, Feil S, et al. cGMP-dependent protein kinase type I is implicated in the regulation of the timing and quality of sleep and wakefulness. PLoS ONE. 2009;4: e4238.
Donlea J, Leahy A, Thimgan MS, et al. Foraging alters resilience/vulnerability to sleep disruption and starvation in Drosophila. Proc Natl Acad Sci U S A. 2012;109:2613–8.
Raizen DM, Zimmerman JE, Maycock MH, et al. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–72.
Kanaya HJ, Park S, Kim JH, et al. A sleep-like state in Hydra unravels conserved sleep mechanisms during the evolutionary development of the central nervous system. Sci Adv. 2020. https://doi.org/10.1126/sciadv.abb9415.
Wong ML, Whelan F, Deloukas P, et al. Phosphodiesterase genes are associated with susceptibility to major depression and antidepressant treatment response. Proc Natl Acad Sci U S A. 2006;103:15124–9.
Lee DI, Zhu G, Sasaki T, et al. Phosphodiesterase 9A controls nitric-oxide-independent cGMP and hypertrophic heart disease. Nature. 2015;519:472–6.
Delhaye S, Bardoni B. Role of phosphodiesterases in the pathophysiology of neurodevelopmental disorders. Mol Psychiatry. 2021. https://doi.org/10.1038/s41380-020-00997-9.
Domek-Lopacinska KU, Strosznajder JB. Cyclic GMP and nitric oxide synthase in aging and Alzheimer’s disease. Mol Neurobiol. 2010;41:129–37.
Harms JF, Menniti FS, Schmidt CJ. Phosphodiesterase 9A in brain regulates cGMP signaling independent of nitric-oxide. Front Neurosci. 2019;13:837.
Orom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58.
Schulz S, Chachami G, Kozaczkiewicz L, et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012;13:930–8.
Bermejo JL, Kabisch M, Dunnebier T, et al. Exploring the association between genetic variation in the SUMO isopeptidase gene USPL1 and breast cancer through integration of data from the population-based GENICA study and external genetic databases. Int J Cancer. 2013;133:362–72.
Toyoda H, Miyagawa T, Koike A, et al. A polymorphism in CCR1/CCR3 is associated with narcolepsy. Brain Behav Immun. 2015;49:148–55.
Acknowledgements
The authors are deeply grateful to all participants in the present study.
Funding
This study was supported by a Practical Research Project for Rare/Intractable Diseases grant from the Japan Agency for Medical Research and Development (AMED), Grants-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (15H04709 and 19H03588), Grants-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (21K07534) and Grants-in-Aid from the Takeda Science Foundation. The funders had no role in study design, data collection, analysis, the decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Dr. Inoue Yuichi has received grants and payment for lectures, including service on speakers’ bureaus, and has provided expert testimony for MSD K.K., Takeda Pharmaceutical Co. Ltd., and Eisai Co. Ltd. Dr. Makoto Honda has received consulting fees from Takeda Pharmaceutical Co. Ltd. The other authors have no competing interests to declare.
Ethics approval
This research involved human participants. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. All individuals provided written informed consent for participation in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tanida, K., Shimada, M., Khor, SS. et al. Genome-wide association study of idiopathic hypersomnia in a Japanese population. Sleep Biol. Rhythms 20, 137–148 (2022). https://doi.org/10.1007/s41105-021-00349-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41105-021-00349-2