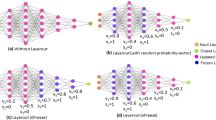
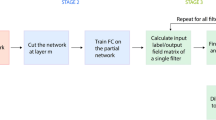
Abstract
Overfitting reduces the generalizability of convolutional neural networks (CNNs). Overfitting is generally detected by comparing the accuracies and losses of the training and validation data, where the validation data are formed from a portion of the training data; however, detection methods are ineffective for pretrained networks distributed without the training data. Thus, in this paper, we propose a method to detect overfitting of CNNs using the trained network weights inspired by the dropout technique. The dropout technique has been employed to prevent CNNs from overfitting, where the neurons in the CNNs are invalidated randomly during their training. It has been hypothesized that this technique prevents CNNs from overfitting by restraining the co-adaptations among neurons, and this hypothesis implies that the overfitting of CNNs results from co-adaptations among neurons and can be detected by investigating the inner representation of CNNs. The proposed persistent homology-based overfitting measure (PHOM) method constructs clique complexes in CNNs using the trained network weights, and the one-dimensional persistent homology investigates co-adaptations among neurons. In addition, we enhance PHOM to normalized PHOM (NPHOM) to mitigate fluctuation in PHOM caused by the difference in network structures. We applied the proposed methods to convolutional neural networks trained for the classification problems on the CIFAR-10, street view house number, Tiny ImageNet, and CIFAR-100 datasets. Experimental results demonstrate that PHOM and NPHOM can indicate the degree of overfitting of CNNs, which suggests that these methods enable us to filter overfitted CNNs without requiring the training data.












Similar content being viewed by others
Availability of data and materials
The datasets are available as referenced in the paper.
Notes
We used the notation from https://towardsdatascience.com/convolutional-neural-networks-mathematics-1beb3e6447c0 with modifications based on our understanding.
The source code and models used in the evaluation can be accessed at https://github.com/satoru-watanabe-aw/phom/.
References
Dictionary, O.: Oxford dictionaries. Language Matters (2014)
Bishop, C.M.: Pattern Recognition and Machine Learning. Springer, Switzerland (2006)
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I., Salakhutdinov, R.: Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15(1), 1929–1958 (2014)
Rieck, B., Togninalli, M., Bock, C., Moor, M., Horn, M., Gumbsch, T., Borgwardt, K.: Neural persistence: a complexity measure for deep neural networks using algebraic topology. In: International Conference on Learning Representations (2018)
Corneanu, C.A., Madadi, M., Escalera, S., Martinez, A.M.: What does it mean to learn in deep networks? and, how does one detect adversarial attacks? In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 4757–4766 (2019)
Corneanu, C.A., Escalera, S., Martinez, A.M.: Computing the testing error without a testing set. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 2677–2685 (2020)
Watanabe, S., Yamana, H.: Topological measurement of deep neural networks using persistent homology. In: ISAIM (2020)
Watanabe, S., Yamana, H.: Topological measurement of deep neural networks using persistent homology. Ann. Math. Artif. Intell. (2021). https://doi.org/10.1007/s10472-021-09761-3
Watanabe, S., Yamana, H.: Deep neural network pruning using persistent homology. In: 2020 IEEE Third International Conference on Artificial Intelligence and Knowledge Engineering (AIKE), pp. 153–156. IEEE (2020)
Otter, N., Porter, M.A., Tillmann, U., Grindrod, P., Harrington, H.A.: A roadmap for the computation of persistent homology. EPJ Data Sci. 6(1), 17 (2017)
Petri, G., Expert, P., Turkheimer, F., Carhart-Harris, R., Nutt, D., Hellyer, P.J., Vaccarino, F.: Homological scaffolds of brain functional networks. J. R. Soc. Interface 11(101), 20140873 (2014)
Cassidy, B., Bowman, F.D., Rae, C., Solo, V.: On the reliability of individual brain activity networks. IEEE Trans. Med. Imaging 37(2), 649–662 (2018). https://doi.org/10.1109/TMI.2017.2774364
Sizemore, A.E., Giusti, C., Kahn, A., Vettel, J.M., Betzel, R.F., Bassett, D.S.: Cliques and cavities in the human connectome. J. Comput. Neurosci. 44(1), 115–145 (2018)
Xia, K., Wei, G.-W.: Persistent homology analysis of protein structure, flexibility, and folding. Int. J. Numer. Methods Biomed. Eng. 30(8), 814–844 (2014)
Gameiro, M., Hiraoka, Y., Izumi, S., Kramar, M., Mischaikow, K., Nanda, V.: A topological measurement of protein compressibility. Jpn. J. Ind. Appl. Math. 32(1), 1–17 (2015)
Kramar, M., Goullet, A., Kondic, L., Mischaikow, K.: Persistence of force networks in compressed granular media. Phys. Rev. E 87(4), 042207 (2013)
Hiraoka, Y., Nakamura, T., Hirata, A., Escolar, E.G., Matsue, K., Nishiura, Y.: Hierarchical structures of amorphous solids characterized by persistent homology. Proc. Natl. Acad. Sci. 113(26), 7035–7040 (2016)
Watanabe, S., Yamana, H.: Overfitting measurement of deep neural networks using no data. In: 2021 IEEE 8th International Conference on Data Science and Advanced Analytics (DSAA), pp. 1–10 (2021). https://doi.org/10.1109/DSAA53316.2021.9564119
Edelsbrunner, H., Harer, J.: Computational Topology: An Introduction. American Mathematical Soc., Providence (2010)
The GUDHI Project: GUDHI User and Reference Manual, GUDHI Editorial Board (2015). http://gudhi.gforge.inria.fr/doc/latest/
Tausz, A., Vejdemo-Johansson, M., Adams, H.: JavaPlex: a research software package for persistent (co)homology. In: Hong, H., Yap, C. (eds.) Proceedings of ICMS 2014. Lecture Notes in Computer Science, vol 8592, pp. 129–136 (2014). Software available at http://appliedtopology.github.io/javaplex/
Edelsbrunner, H., Morozov, D.: Persistent homology: theory and practice. Technical report, Lawrence Berkeley National Lab. (LBNL), Berkeley, CA (United States) (2012)
Edelsbrunner, H., Letscher, D., Zomorodian, A.: Topological persistence and simplification. In: Proceedings 41st Annual Symposium on Foundations of Computer Science, pp. 454–463. IEEE (2000)
Masulli, P., Villa, A.E.: The topology of the directed clique complex as a network invariant. Springerplus 5(1), 388 (2016)
Montavon, G., Lapuschkin, S., Binder, A., Samek, W., Müller, K.-R.: Explaining nonlinear classification decisions with deep Taylor decomposition. Pattern Recogn. 65, 211–222 (2017)
Virtanen, P., Gommers, R., Oliphant, T.E., Haberland, M., Reddy, T., Cournapeau, D., Burovski, E., Peterson, P., Weckesser, W., Bright, J., et al.: Scipy 1.0: fundamental algorithms for scientific computing in python. Nat. Methods 17(3), 261–272 (2020)
Krizhevsky, A., Hinton, G., et al.: Learning multiple layers of features from tiny images (2009)
Netzer, Y., Wang, T., Coates, A., Bissacco, A., Wu, B., Ng, A.Y.: Reading digits in natural images with unsupervised feature learning (2011)
Le, Y., Yang, X.: Tiny imagenet visual recognition challenge. CS 231N 7(7), 3 (2015)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. In: Bengio, Y., LeCun, Y. (eds.) 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, May 7–9, 2015, Conference Track Proceedings (2015)
Gulli, A., Pal, S.: Deep Learning with Keras. Packt Publishing Ltd., Birmingham (2017)
Yosinski, J., Clune, J., Bengio, Y., Lipson, H.: How transferable are features in deep neural networks? Adv. Neural Inf. Process. Syst. 27, 3320–3328 (2014)
Chollet, F., et al.: Deep Learning with Python, vol. 361. Manning New York, New York (2018)
Mishra, R., Gupta, H.P., Dutta, T.: A survey on deep neural network compression: Challenges, overview, and solutions. arXiv preprint arXiv:2010.03954 (2020)
Blalock, D., Gonzalez Ortiz, J.J., Frankle, J., Guttag, J.: What is the state of neural network pruning? In: Dhillon, I., Papailiopoulos, D., Sze, V. (eds.) Proceedings of Machine Learning and Systems, vol. 2, pp. 129–146 (2020)
Edelsbrunner, H., Harer, J., et al.: Persistent homology—a survey. Contemp. Math. 453, 257–282 (2008)
Werpachowski, R., György, A., Szepesvari, C.: Detecting overfitting via adversarial examples. Adv. Neural Inf. Process. Syst. 32, 7858–7868 (2019)
Grosse, K., Lee, T., Park, Y., Backes, M., Molloy, I.M.: A new measure for overfitting and its implications for backdooring of deep learning. CoRR arXiv: 2006.06721 (2020)
Raghu, M., Gilmer, J., Yosinski, J., Sohl-Dickstein, J.: SVCCA: singular vector canonical correlation analysis for deep learning dynamics and interpretability. In: Advances in Neural Information Processing Systems, pp. 6076–6085 (2017)
Morcos, A., Raghu, M., Bengio, S.: Insights on representational similarity in neural networks with canonical correlation. In: Advances in Neural Information Processing Systems, pp. 5727–5736 ( 2018)
Kornblith, S., Norouzi, M., Lee, H., Hinton, G.: Similarity of neural network representations revisited. In: International Conference on Machine Learning, pp. 3519–3529 (2019)
Acknowledgements
We are grateful to Hitachi, Ltd. for the tuition subsidy. The founder had no role in both study design and technical investigation in this paper.
Funding
This research has not received any funding supports.
Author information
Authors and Affiliations
Contributions
The authors proposed a persistent homology-based overfitting measure that detects the overfitting of convolutional neural networks without relying on the training data.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest and competing interests about this paper.
Code availability
The source codes are available as referenced in the paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Watanabe, S., Yamana, H. Overfitting measurement of convolutional neural networks using trained network weights. Int J Data Sci Anal 14, 261–278 (2022). https://doi.org/10.1007/s41060-022-00332-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41060-022-00332-1