Abstract
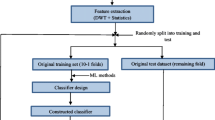
For evaluating performance of nonlinear features and iterative and non-iterative classification algorithms (i.e. kernel support vector machine (KSVM), random forest (RaF), least squares SVM (LS-SVM) and multi-surface proximal SVM based oblique RaF (ORaF) for ECG quality assessment we compared the four algorithms on 7 feature schemes yielded from 27 linear and nonlinear features including four features derived from a new encoding Lempel–Ziv complexity (ELZC) and the other 26 features. Seven feature schemes include the first scheme consisting of 7 waveform features, the second consisting of 15 waveform and frequency features, the third consisting of 19 waveform, frequency and approximate entropy (ApEn) features, the fourth consisting of 19 waveform, frequency and permutation entropy (PE) features, the fifth consisting of 19 waveform, frequency and ELZC features, the sixth consisting of 23 waveform, frequency, PE and ELZC features, and the last consisting of all 27 features. Up to 1500 mobile ECG recordings from the Physionet/Computing in Cardiology Challenge 2011 were employed in this study. Three indices i.e., sensitivity (Se), specificity (Sp) and accuracy (Acc), were used for evaluating performances of the classifiers on the seven feature schemes, respectively. The experiment results indicated PE and ELZC can help to improve performance of the aforementioned four classifiers for assessing ECG quality. Using all features except ApEn features obtained the best performances for each classifier. For this sixth scheme, the LS-SVM yielded the highest Acc of 92.20% on hidden test data, as well as a relatively high Acc of 93.60% on training data. Compared with the other classifiers, the LS-SVM classifier also demonstrated the superior generalization ability.



Similar content being viewed by others
References
Li, Q., & Clifford, G. D. (2012). Signal quality and data fusion for false alarm reduction in the intensive care unit. Journal of Electrocardiology, 45, 596–603.
Zhang, Y. T., Liu, C. Y., Wei, S. S., Wei, C. Z., & Liu, F. F. (2014). ECG quality assessment based on a kernel support vector machine and genetic algorithm with a feature matrix. Journal of Zhejiang University Science C, 15, 564–573.
Clifford, G. D., Behar, J., Li, Q., & Rezek, I. (2012). Signal quality indices and data fusion for determining clinical acceptability of electrocardiograms. Physiological Measurement, 33, 1419–1433.
Li, Q., & Clifford, G. D. (2012). Dynamic time warping and machine learning for signal quality assessment of pulsatile signals. Physiological Measurement, 33, 1491–1501.
Orphanidou, C., & Drobnjak, I. (2017). Quality assessment of ambulatory ECG using wavelet entropy of the HRV signal. IEEE Journal of Biomedical and Health Informatics, 21, 1216–1223.
Langley, P., Di Marco, L. Y., King, S., Duncan, D., Di Maria, C., Duan, W., Bojarnejad, M., Zheng, D., Allen, J., & Murray, A. (2011). An algorithm for assessment of quality of ECGs acquired via mobile telephones. In Computing in Cardiology, IEEE, Hangzhou (Vol. 38, pp. 281–284).
Johannesen, L. (2011). Assessment of ECG quality on an Android platform. In Computing in Cardiology, IEEE, Hangzhou (Vol. 38, pp. 433–436).
Zaunseder, S., Huhle, R., & Malberg, H. (2011). Assessing the usability of ECG by ensemble decision trees. In Computing in Cardiology, IEEE, Hangzhou (Vol. 38, pp. 277–280).
Chen, Y., & Yang, H. (2012). Self-organized neural network for the quality control of 12-lead ECG signals. Physiological Measurement, 33, 1399–1418.
Kužílek, J., Huptych, M., Chudáček, V., Spilka1, J., Lhotská, L. (2011). Data driven approach to ECG signal quality assessment using multistep SVM classification. In Computing in Cardiology, IEEE, Hangzhou (Vol. 38, pp. 453–455).
Li, Q., Rajagopalan, C., & Clifford, G. D. (2014). A machine learning approach to multi-level ECG signal quality classification. Computer Methods and Programs in Biomedicine, 117, 435–447.
Y. Zhang, S. Wei, L. Zhang and C. Liu, A signal quality assessment method for mobile ECG using multiple features and fuzzy support vector machine. In Natural Computation, Fuzzy Systems and Knowledge Discovery (ICNC-FSKD), 12th International Conference on (pp. 966–971). IEEE. 2016.
Suykens, J. A. K., Van Gestel, T., De Brabanter, J., De Moor, B., & Vandewalle, J. (2002). Least squares support vector machines. Singapore: World Scientific.
Zhang, L., & Suganthan, P. N. (2014). Random forests with ensemble of feature spaces. Pattern Recognition, 47, 3429–3437.
Zhang, L., & Suganthan, P. N. (2015). Oblique decision tree ensemble via multisurface proximal support vector machine. IEEE Transactions on Cybernetics, 45, 2165–2176.
Zhang, L., & Suganthan, P. N. (2016). A survey of randomized algorithms for training neural networks. Information Sciences, 364, 146–155.
Zhang, L., & Suganthan, P. N. (2015). A comprehensive evaluation of random vector functional link networks. Information Sciences, 367, 1094–1105.
Mohapatra, P., Chakravarty, S., & Dash, P. K. (2016). Microarray medical data classification using kernel ridge regression and modified cat swarm optimization based gene selection system. Swarm and Evolutionary Computation, 28, 144–160.
Moody, G. B. (2011). Physionet/computing in cardiology challenge 2011, July 2011. http://physionet.org/challenge/2011. Accessed 5 Oct 2017.
Shetty, P., & Bhat, S. (2014). Analysis of various filter configurations on noise reduction in ECG waveform. International Journal of Computing, Communications & Instrumentation Engg, 1, 88–91.
Richman, J. S., & Moorman, J. R. (2000). Physiological time-series analysis using approximate entropy and sample entropy. American Journal of Physiology-Heart and Circulatory Physiology., 278, H2039–H2049.
Bandt, C., & Pompe, B. (2002). Permutation entropy: A natural complexity measure for time series. Physical Review Letters, 88, 174102.
Zhang, Y. T., Wei, S. S., Liu, H., Zhao, L. N., & Liu, C. Y. (2016). A novel encoding Lempel-Ziv complexity algorithm for quantifying the irregularity of physiological time series. Computer Methods and Programs in Biomedicine, 133, 7–15.
Zhang, C. S. (2000). Optimization of Kernel function parameters SVM based on the GA. Advanced Materials Research, 433, 4124–4128.
Zhang, C. S. (2000). Optimization of Kernel function parameters SVM based on the GA. Advanced Materials Research, 433, 4124–4128.
Wang, X., Yang, C., Qin, B., & Gui, W. (2005). Parameter selection of support vector regression based on hybrid optimization algorithm and its application. Journal of Control Theory and Applications, 3, 371–376.
Masetic, Z., & Subasi, A. (2016). Congestive heart failure detection using random forest classifier. Computer Methods and Programs in Biomedicine, 130, 54–64.
Breiman, L. (2001). Random forests. Machine Learning, 45, 5–32.
Khazaee, A., & Ebrahimzadeh, A. (2010). Classification of electrocardiogram signals with support vector machines and genetic algorithms using power spectral features. Biomedical Signal Processing and Control, 5, 252–263.
Mangasarian, O. L., & Wild, E. W. (2006). Multisurface proximal support vector machine classification via generalized eigenvalues. IEEE Transactions on Pattern Analysis and Machine Intelligence, 28, 69–74.
Wang, L., Wang, Y., & Chang, Q. (2016). Feature selection methods for big data bioinformatics: A survey from the search perspective. Methods, 111, 21–31.
Acknowledgment
We thank the MIT-BIH database for providing the invaluable data used in this study. We also thank Dr. Le Zhang and Prof. Ponnuthurai N. Suganthan, because they provide the public code of the ORaF based on multi-surface proximal support vector machine. This work was supported by Postdoctoral Science Foundation of China (No. 2017M612280), National Natural Science Foundation of China (No. 61473174), and Natural Science Foundation of Shandong Province, China (Nos. ZR2015AM015 and ZR2017MA046).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Rights and permissions
About this article
Cite this article
Zhang, Y., Wei, S., Zhang, L. et al. Comparing the Performance of Random Forest, SVM and Their Variants for ECG Quality Assessment Combined with Nonlinear Features. J. Med. Biol. Eng. 39, 381–392 (2019). https://doi.org/10.1007/s40846-018-0411-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40846-018-0411-0