Abstract
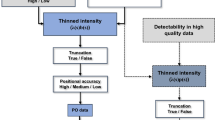
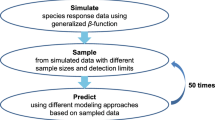
Species distribution models (SDMs) are crucial in ecology, conservation, and ecosystem management. Numerous SDMs have been developed over time, and studies have shown that these tools can be affected by a range of factors, such as data type, spatial resolution, number of explanatory variables, sample characteristics, and collinearity between environmental variables. New SDMs have often been developed to address some of these issues. Understanding the performance of new statistical tools is crucial for researchers in various fields. Thus, we assessed the predictive ability of the Poisson point process and log Gaussian Cox process models, considered as new SDMs, by simulating two factors, spatial resolution and imperfect detection, which are likely to have significant effects on SDMs, and considering Gabon as the study area. The observed model performance metrics, such as the Area Under the Receiver Operating Characteristic Curve (AUC), Mean Absolute Error (MAE), and Pearson correlation (CORR) between the true and predicted intensities, were used to evaluate the predictive performance of these models. The results showed that, although most of these models failed to estimate the intercept \({\alpha }_{0}\) and covariate coefficients (\({\varvec{\beta }}_{x}{\textbf {x}}(z)\)) correctly, they at least had the merit of demonstrated good performance (AUC more than 70%, CORR more than 67%, and MAE less than 0.61%). However, the spatial resolution of the environmental variables and imperfect detection of simulated species occurrences significantly affected the predictive performance of the two models (P < .0001). This study offers important insights for ecologist modellers, environmentalists, and conservators.










Similar content being viewed by others
Data availability
The data supporting the findings of this study are available on request from the corresponding author.
References
Abrego N, Ovaskainen O (2023) Evaluating the predictive performance of presence-absence models: why can the same model appear excellent or poor? Ecol Evolut 13(12):e10784. https://doi.org/10.1002/ece3.10784
Anderson RP, Peterson AT, Gómez-Laverde M (2002) Using niche-based GIS modeling to test geographic predictions of competitive exclusion and competitive release in south American pocket mice. Oikos 98(1):3–16
Austin M (2007) Species distribution models and ecological theory: a critical assessment and some possible new approaches. Ecol Model 200(1):1–19. https://doi.org/10.1016/j.ecolmodel.2006.07.005
Baddeley A, Rubak E, Turner R (2016) Spatial Point Patterns: Methodology and Applications with R. Chapman & Hall/CRC
Bahn V, McGill BJ (2013) Testing the predictive performance of distribution models. Oikos 122(3):321–331. https://doi.org/10.1111/j.1600-0706.2012.00299.x
Baker DJ, Maclean IMD, Goodall M et al (2021) Species distribution modelling is needed to support ecological impact assessments. J Appl Ecol 58(1):21–26. https://doi.org/10.1111/1365-2664.13782
Banerjee (2004) Hierarchical modeling and analysis for spatial data. Chapman and Hall/CRC Press
Besag J (1975) Statistical analysis of non-lattice data. J R Stat Soc Ser D (Statist) 24(3):179–195
Buri A, Cianfrani C, Pinto-Figueroa E et al (2017) Soil factors improve predictions of plant species distribution in a mountain environment. Prog Phys Geogr Earth Environ 41(6):703–722. https://doi.org/10.1177/0309133317738162
Chauvier Y, Descombes P, Guéguen M et al (2022) Resolution in species distribution models shapes spatial patterns of plant multifaceted diversity. Ecography 10:e05973
Cressie NAC (1993) Statistics for spatial data. Wiley, Hoboken
De Marco P, Nóbrega CC (2018) Evaluating collinearity effects on species distribution models: an approach based on virtual species simulation. PLOS One 13(9):e0202403. https://doi.org/10.1371/journal.pone.0202403
Dorazio RM (2014) Accounting for imperfect detection and survey bias in statistical analysis of presence-only data. Global Ecol Biogeogr 23(12):1472–1484
Elith J, Leathwick JR (2009) Species distribution models: ecological explanation and prediction across space and time. Annu Rev Ecol Evolut System 40(1):677–697. https://doi.org/10.1146/annurev.ecolsys.110308.120159
Elith JH, Graham CP, Anderson R et al (2006) Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29(2):129–151
Elith J, Kearney M, Phillips S (2010) The art of modelling range-shifting species. Methods Ecol Evolut 1(4):330–342. https://doi.org/10.1111/j.2041-210X.2010.00036.x
Fao (2012) Harmonized world soil database (version 1.2). FAO, Rome, Italy and IIASA, Laxenburg, Austria
Fei S, Yu F (2016) Quality of presence data determines species distribution model performance: a novel index to evaluate data quality. Landsc Ecol 31(1):31–42. https://doi.org/10.1007/s10980-015-0272-7
Fick SE, Hijmans RJ (2017) WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol 37(12):4302–4315
Fithian W, Hastie T (2013) Finite-sample equivalence in statistical models for presence-only data. Ann Appl Stat 7(4):1917. https://doi.org/10.1214/13-aoas667
Fithian W, Elith J, Hastie T et al (2015) Bias correction in species distribution models: pooling survey and collection data for multiple species. Methods Ecol Evolut 6(4):424–438. https://doi.org/10.1111/2041-210x.12242
Fletcher RJ, McCleery RA, Greene DU et al (2016) Integrated models that unite local and regional data reveal larger-scale environmental relationships and improve predictions of species distributions. Landsc Ecol 31(6):1369–1382
Fox J, Weisberg S, Adler D, et al (2012) Package ‘car’. R Foundation for Statistical Computing 16, Vienna
Franklin J (2009) Mapping species distributions: spatial inference and prediction. Cambridge University Press
Graham CH, Elith J, Hijmans RJ et al (2008) The influence of spatial errors in species occurrence data used in distribution models. J Appl Ecol 45(1):239–247
Guélat J, Kéry M (2018) Effects of spatial autocorrelation and imperfect detection on species distribution models. Methods Ecol Evolut 9(6):1614–1625
Guisan A, Thuiller W (2005) Predicting species distribution: offering more than simple habitat models. Ecol Lett 8(9):993–1009. https://doi.org/10.1111/j.1461-0248.2005.00792.x
Gábor L, Moudrý V, Barták V et al (2020) How do species and data characteristics affect species distribution models and when to use environmental filtering? Int J Geograph Inform Sci 34(8):1567–1584. https://doi.org/10.1080/13658816.2019.1615070
Hallgren W, Santana F, Low-Choy S et al (2019) Species distribution models can be highly sensitive to algorithm configuration. Ecol Model 408(108):719. https://doi.org/10.1016/j.ecolmodel.2019.108719
Hijmans RJ, Cruz M, Rojas E et al (2001) DIVA-GIS, version 1.4. a geographic information system for the management and analysis of genetic resources data. Plant Gen Resour Newslett 127:15–19
Holdridge LR, Grenke WC (1971) Forest environments in tropical life zones: a pilot study. Forest environments in tropical life zones: a pilot study
Hothorn T, Bretz F, Westfall P et al (2016) Package ‘multcomp’. Simultaneous inference in general parametric models Project for Statistical Computing, Vienna, Austria
Humphreys JM, Elsner JB, Jagger TH et al (2017) A bayesian geostatistical approach to modeling global distributions of lygodium microphyllum under projected climate warming. Ecol Model 363:192–206
Huston MA (2005) Introductory essay: critical issues for improving predictions. In: Predicting species occurrences: issues of accuracy and scale
Irving K, Jähnig SC, Kuemmerlen M (2020) Identifying and applying an optimum set of environmental variables in species distribution models. Inland Waters 10(1):11–28. https://doi.org/10.1080/20442041.2019.1653111
Isaac NJ, Jarzyna MA, Keil P, et al (2019) Data integration for large-scale models of species distributions. Trends Ecol Evolut
Isaac NJB, Pocock MJO (2015) Bias and information in biological records: bias and information in biological records. Biol J Linnean Soc 115(3):522–531. https://doi.org/10.1111/bij.12532
Johnson DS, Hooten MB, Kuhn CE (2013) Estimating animal resource selection from telemetry data using point process models. J Anim Ecol 82(6):1155–1164
Khan AM, Li Q, Saqib Z, et al (2022) MaxEnt modelling and impact of climate change on habitat suitability variations of economically important chilgoza pine (Pinus gerardiana wall.) in south Asia. Forests 13(5):715. https://www.mdpi.com/1999-4907/13/5/715
Koshkina V, Wang Y, Gordon A et al (2017) Integrated species distribution models: combining presence-background data and site-occupancy data with imperfect detection. Methods Ecol Evolut 8(4):420–430
Lavancier F, Poinas A, Waagepetersen R (2021) Adaptive estimating function inference for nonstationary determinantal point processes. Scand J Stat 48(1):87–107
Levin SA (1992) The problem of pattern and scale in ecology: the robert h MacArthur award lecture. Ecology 73(6):1943–1967
Liu C, Wolter C, Xian W et al (2020) Species distribution models have limited spatial transferability for invasive species. Ecol Lett 23(11):1682–1692. https://doi.org/10.1111/ele.13577
Loiselle BA, Jørgensen PM, Consiglio T et al (2008) Predicting species distributions from herbarium collections: does climate bias in collection sampling influence model outcomes? J Biogeogr 35(1):105–116
Mandrekar JN (2010) Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol 5(9):1315–1316
Meineri E, Hylander K (2017) Fine-grain, large-domain climate models based on climate station and comprehensive topographic information improve microrefugia detection. Ecography 40(8):1003–1013. https://doi.org/10.1111/ecog.02494
Mengersen K, Peterson EE, Clifford S et al (2017) Modelling imperfect presence data obtained by citizen science. Environmetrics 28(5):e2446
Merow C, Wilson AM, Jetz W (2017) Integrating occurrence data and expert maps for improved species range predictions. Global Ecol Biogeogr 26(2):243–258
Miller J (2010) Species distribution modeling: species distribution modeling. Geogr Compass 4(6):490–509. https://doi.org/10.1111/j.1749-8198.2010.00351.x
Moraga P, Cano J, Baggaley RF et al (2015) Modelling the distribution and transmission intensity of lymphatic filariasis in sub-saharan africa prior to scaling up interventions: integrated use of geostatistical and mathematical modelling. Parasit Vect 8(1):560
Moura Júnior EGd, Nascimento FAOd, Lemos Filho JPd et al (2021) Limnological layers improve species distribution modeling of aquatic macrophytes at fine-spatial resolution. Acta Bot Brasil 35:9–16
Naimi B, Araújo MB (2016) sdm: a reproducible and extensible r platform for species distribution modelling. Ecography 39(4):368–375
Norberg A, Abrego N, Blanchet FG, et al. (2019) A comprehensive evaluation of predictive performance of 33 species distribution models at species and community levels. Ecol Monogr 89(3). https://doi.org/10.1002/ecm.1370
Osborne J (2010) Improving your data transformations: applying the box-cox transformation. Pract Assess Res Evaluat 15(1):12
Pearce JL, Boyce MS (2006) Modelling distribution and abundance with presence-only data. J Appl Ecol 43(3):405–412. https://doi.org/10.1111/j.1365-2664.2005.01112.x
Peel SL, Hill NA, Foster SD et al (2019) Reliable species distributions are obtainable with sparse, patchy and biased data by leveraging over species and data types. Methods Ecol Evolut 10(7):1002–1014. https://doi.org/10.1111/2041-210x.13196
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190(3):231–259. https://doi.org/10.1016/j.ecolmodel.2005.03.026
Phillips SJ, Anderson RP, Dudík M et al (2017) Opening the black box: an open-source release of maxent. Ecography 40(7):887–893. https://doi.org/10.1111/ecog.03049
Rajala T (2014) A note on bayesian logistic regression for spatial exponential family gibbs point processes. arXiv preprint arXiv:1411.0539
Rajala T, Penttinen A (2014) Bayesian analysis of a gibbs hard-core point pattern model with varying repulsion range. Comput Stati Data Anal 71:530–541
Randin CF, Engler R, Normand S et al (2009) Climate change and plant distribution: local models predict high-elevation persistence. Global Change Biol 15(6):1557–1569
Renner IW, Elith J, Baddeley A et al (2015) Point process models for presence-only analysis. Methods Ecol Evolut 6(4):366–379. https://doi.org/10.1111/2041-210x.12352
Reutter BA, Helfer V, Hirzel AH et al (2003) Modelling habitat-suitability using museum collections: an example with three sympatric Apodemus species from the alps. J Biogeogr 30(4):581–590. https://doi.org/10.1046/j.1365-2699.2003.00855.x
Rose JP, Halstead BJ, Fisher RN (2020) Integrating multiple data sources and multi-scale land-cover data to model the distribution of a declining amphibian. Biol Conserv 241(108):374. https://doi.org/10.1016/j.biocon.2019.108374
Shabani F, Kumar L, Ahmadi M (2016) A comparison of absolute performance of different correlative and mechanistic species distribution models in an independent area. Ecol Evolut 6(16):5973–5986. https://doi.org/10.1002/ece3.2332
Simmonds EG, Jarvis SG, Henrys PA et al (2020) Is more data always better? A simulation study of benefits and limitations of integrated distribution models. Ecography 43(10):1413–1422. https://doi.org/10.1111/ecog.05146
Srivastava V, Lafond V, Griess VC (2019) Species distribution models (sdm): applications, benefits and challenges in invasive species management. CABI Rev:1–13. https://doi.org/10.1079/PAVSNNR201914020
Stralberg D, Matsuoka SM, Hamann A et al (2015) Projecting boreal bird responses to climate change: the signal exceeds the noise. Ecol Appl 25(1):52–69. https://doi.org/10.1890/13-2289.1
Støa B, Halvorsen R, Mazzoni S et al (2018) Sampling bias in presence-only data used for species distribution modelling: theory and methods for detecting sample bias and its effects on models. Sommerfeltia 38(1):1–53
Tanaka U, Ogata Y, Stoyan D (2008) Parameter estimation and model selection for Neyman–Scott point processes. Biomet J 50(1):43–57. https://doi.org/10.1002/bimj.200610339
Team RC (2022) R: a language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria
Thompson CG, Kim RS, Aloe AM et al (2017) Extracting the variance inflation factor and other multicollinearity diagnostics from typical regression results. Basic Appl Soc Psychol 39(2):81–90
Václavík T, Kupfer JA, Meentemeyer RK (2012) Accounting for multi-scale spatial autocorrelation improves performance of invasive species distribution modelling (iSDM): Multi-scale spatial autocorrelation and invasive species distribution models. J Biogeogr 39(1):42–55. https://doi.org/10.1111/j.1365-2699.2011.02589.x
Valavi R, Shafizadeh-Moghadam H, Matkan A et al (2019) Modelling climate change effects on Zagros forests in Iran using individual and ensemble forecasting approaches. Theor Appl Climatol 137(1):1015–1025 (Publisher: Springer)
Valavi R, Guillera-Arroita G, Lahoz-Monfort JJ et al (2021) Predictive performance of presence-only species distribution models: a benchmark study with reproducible code. Ecol Monogr 1(e01):486
van Proosdij AS, Sosef MS, Wieringa JJ et al (2016) Minimum required number of specimen records to develop accurate species distribution models. Ecography 39(6):542–552
Walsh MG (2015) Mapping the risk of Nipah virus spillover into human populations in south and southeast Asia. Trans R Soc Trop Med Hygiene 109(9):563–571
Ward G, Hastie T, Barry S et al (2009) Presence-only data and the EM algorithm. Biometrics 65(2):554–563
Warton DI, Shepherd LC (2010) Poisson point process models solve the “pseudo-absence problem’’ for presence-only data in ecology. Ann Appl Stat 4(3):1383–1402. https://doi.org/10.1214/10-aoas331
Warton DI, Renner IW, Ramp D (2013) Model-based control of observer bias for the analysis of presence-only data in ecology. PLoS One 8(11):e79168. https://doi.org/10.1371/journal.pone.0079168
Williams JN, Seo C, Thorne J et al (2009) Using species distribution models to predict new occurrences for rare plants. Divers Distrib 15(4):565–576. https://doi.org/10.1111/j.1472-4642.2009.00567.x
Wisz MS, Hijmans RJ, Li J et al (2008) Effects of sample size on the performance of species distribution models. Diver Distrib 14(5):763–773
Acknowledgements
We are grateful to the reviewers and editors for their constructive feedback, insightful comments, and the time they devoted to reviewing this manuscript.
Funding
This work was carried out as part of the “Ph.D. In-Country/In-Region Scholarship Programme FSA/UAC, 2020 (Grant number 91786077 to JABB)” of the German Federal Ministry of Economic Cooperation and Development (BMZ). JABB is grateful to the German Academic Exchange Service (DAAD) for funding his Ph.D. studies.
Author information
Authors and Affiliations
Contributions
JABB: conceptualisation, methodology, data simulation, data analysis, and writing the initial draft; MLZ: data simulation; ABF: Validation, supervision, and writing revisions; RLGK: Validation, supervision, and writing revisions; DAAD: Funding acquisition.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no Conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bourobou, J.A.B., Zinzinhedo, M.L., Fandohan, A.B. et al. Evaluating spatial resolution and imperfect detection effects on the predictive performance of inhomogeneous spatial point process models trained with simulated presence-only data. Model. Earth Syst. Environ. (2024). https://doi.org/10.1007/s40808-024-02017-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40808-024-02017-z