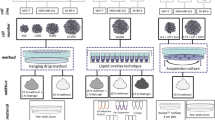
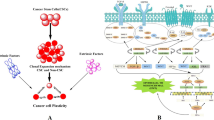
Abstract
Purpose of Review
Tumors consist of heterogeneous cell types, which present challenges to effective therapeutics. This review intends to explore existing mathematical models to better understand the influence of these different types of cells on tumor growth and survival.
Recent Findings
Cancer stem cells are often the instigator of tumor development and drug resistance. The replenishment of the stem cells by other stem cells and progenitor cells impacts the efficacy of treatments. Multiple treatments are required to attack the multiple tumor cell types and induce remission. Mathematical models can be used to explore the behavior of these heterogeneous tumor cells, as well as predict the long-term efficacy of different therapies.
Summary
Cell division plays an integral role in the development of tumors. While mathematical models are generally robust, they must be updated frequently to accommodate the brisk pace of biological advances. Usable data to inform the models is scarce calling for better collaboration between these sciences to help advance the field of cancer therapeutics.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–8. https://doi.org/10.1073/pnas.0530291100.
Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17(24):3029–35. https://doi.org/10.1101/gad.1143403.
Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–11. https://doi.org/10.1038/35102167.
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. https://doi.org/10.1038/nature05236.
Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–7. https://doi.org/10.1158/0008-5472.CAN-06-2030.
Sellheyer K. Basal cell carcinoma: cell of origin, cancer stem cell hypothesis and stem cell markers. Br J Dermatol. 2011;164(4):696–711. https://doi.org/10.1111/j.1365-2133.2010.10158.x.
Wang R, Chadalavada K, Wilshire J, Kowalik U, Hovinga KE, Geber A, et al. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468(7325):829–33. https://doi.org/10.1038/nature09624.
Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13(2):153–66. https://doi.org/10.1016/j.ccr.2008.01.013.
Zhang P, Zuo H, Ozaki T, Nakagomi N, Kakudo K. Cancer stem cell hypothesis in thyroid cancer. Pathol Int. 2006;56(9):485–9. https://doi.org/10.1111/j.1440-1827.2006.01995.x.
Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138(5):822–9. https://doi.org/10.1016/j.cell.2009.08.017.
Natarajan TG, Ganesan N, Fitzgerald KT. Cancer stem cells and markers: new model of tumorigenesis with therapeutic implications. Cancer Biomark. 2010;9(1–6):65–99. https://doi.org/10.3233/CBM-2011-0173.
Wang T, Shigdar S, Gantier MP, Hou Y, Wang L, Li Y, et al. Cancer stem cell targeted therapy: progress amid controversies. Oncotarget. 2015;6(42):44191–206. https://doi.org/10.18632/oncotarget.6176.
Aghaalikhani N, Rashtchizadeh N, Shadpour P, Allameh A, Mahmoodi M. Cancer stem cells as a therapeutic target in bladder cancer. J Cell Physiol. 2019;234(4):3197–206. https://doi.org/10.1002/jcp.26916.
Parizadeh SM, Jafarzadeh-Esfehani R, Hassanian SM, Parizadeh SMR, Vojdani S, Ghandehari M, et al. Targeting cancer stem cells as therapeutic approach in the treatment of colorectal cancer. Int J Biochem Cell Biol. 2019;110:75–83. https://doi.org/10.1016/j.biocel.2019.02.010.
Visweswaran M, Arfuso F, Warrier S, Dharmarajan A. Aberrant lipid metabolism as an emerging therapeutic strategy to target cancer stem cells. Stem Cells. 2020;38(1):6–14. https://doi.org/10.1002/stem.3101.
Xu J, Liao K, Zhou W. Exosomes regulate the transformation of cancer cells in cancer stem cell homeostasis. Stem Cells Int. 2018;2018:4837370. https://doi.org/10.1155/2018/4837370.
Rhodes A, Hillen T. Mathematical modeling of the role of survivin on dedifferentiation and radioresistance in cancer. Bull Math Biol. 2016;78(6):1162–88. https://doi.org/10.1007/s11538-016-0177-x.
Marzagalli M, Fontana F, Raimondi M, Limonta P. Cancer stem cells-key players in tumor relapse. Cancers (Basel). 2021;13(3). https://doi.org/10.3390/cancers13030376.
Ganguly R, Puri IK. Mathematical model for the cancer stem cell hypothesis. Cell Prolif. 2006;39(1):3–14. https://doi.org/10.1111/j.1365-2184.2006.00369.x.
Ganguly R, Puri IK. Mathematical model for chemotherapeutic drug efficacy in arresting tumour growth based on the cancer stem cell hypothesis. Cell Prolif. 2007;40(3):338–54. https://doi.org/10.1111/j.1365-2184.2007.00434.x.
Leder K, Holland EC, Michor F. The therapeutic implications of plasticity of the cancer stem cell phenotype. PLoS One. 2010;5(12): e14366. https://doi.org/10.1371/journal.pone.0014366.
Sehl ME, Sinsheimer JS, Zhou H, Lange KL. Differential destruction of stem cells: implications for targeted cancer stem cell therapy. Cancer Res. 2009;69(24):9481–9. https://doi.org/10.1158/0008-5472.Can-09-2070.
Zhu X, Zhou X, Lewis MT, Xia L, Wong S. Cancer stem cell, niche and EGFR decide tumor development and treatment response: a bio-computational simulation study. J Theor Biol. 2011;269(1):138–49. https://doi.org/10.1016/j.jtbi.2010.10.016.
Busse JE, Gwiazda P, Marciniak-Czochra A. Mass concentration in a nonlocal model of clonal selection. J Math Biol. 2016;73(4):1001–33. https://doi.org/10.1007/s00285-016-0979-3.
Enderling H, Hlatky L, Hahnfeldt P. Cancer Stem Cells: A minor cancer subpopulation that redefines global cancer features. Fronti Oncol. 2013;3(76). https://doi.org/10.3389/fonc.2013.00076.
Liu X, Johnson S, Liu S, Kanojia D, Yue W, Singh UP, et al. Nonlinear growth kinetics of breast cancer stem cells: implications for cancer stem cell targeted therapy. Sci Rep. 2013;3:2473. https://doi.org/10.1038/srep02473.
Molina-Pena R, Alvarez MM. A simple mathematical model based on the cancer stem cell hypothesis suggests kinetic commonalities in solid tumor growth. PLoS One. 2012;7(2): e26233. https://doi.org/10.1371/journal.pone.0026233.
• Molina-Pena R, Tudon-Martinez JC, Aquines-Gutierrez O. A mathematical model of average dynamics in a stem cell hierarchy suggests the combinatorial targeting of cancer stem cells and progenitor cells as a potential strategy against tumor growth. Cancers (Basel). 2020;12(9). https://doi.org/10.3390/cancers12092590. This article argues that a hierarchical model for cell type is relevant because progenitor cells play an integral role in the forming and sustaining tumors. The role of different cell types is examined as radiation therapy is given.
Vainstein V, Kirnasovsky OU, Kogan Y, Agur Z. Strategies for cancer stem cell elimination: insights from mathematical modeling. J Theor Biol. 2012;298:32–41. https://doi.org/10.1016/j.jtbi.2011.12.016.
Weekes SL, Barker B, Bober S, Cisneros K, Cline J, Thompson A, et al. A multicompartment mathematical model of cancer stem cell-driven tumor growth dynamics. Bull Math Biol. 2014;76(7):1762–82. https://doi.org/10.1007/s11538-014-9976-0.
Yan H, Konstorum A, Lowengrub JS. Three-dimensional spatiotemporal modeling of colon cancer organoids reveals that multimodal control of stem cell self-renewal is a critical determinant of size and shape in early stages of tumor growth. Bull Math Biol. 2018;80(5):1404–33. https://doi.org/10.1007/s11538-017-0294-1.
Lionetti MC, Cola F, Chepizhko O, Fumagalli MR, Font-Clos F, Ravasio R, et al. MicroRNA-222 regulates melanoma plasticity. J Clin Med. 2020;9(8):2573.
Sottoriva A, Verhoeff JJ, Borovski T, McWeeney SK, Naumov L, Medema JP, et al. Cancer stem cell tumor model reveals invasive morphology and increased phenotypical heterogeneity. Cancer Res. 2010;70(1):46–56. https://doi.org/10.1158/0008-5472.Can-09-3663.
Tonekaboni SAM, Dhawan A, Kohandel M. Mathematical modelling of plasticity and phenotype switching in cancer cell populations. Math Biosci. 2017;283:30–7. https://doi.org/10.1016/j.mbs.2016.11.008.
Zapperi S, La Porta CA. Do cancer cells undergo phenotypic switching? The case for imperfect cancer stem cell markers. Sci Rep. 2012;2:441. https://doi.org/10.1038/srep00441.
Afenya EK, Ouifki R, Camara BI, Mundle SD. Mathematical modeling of bone marrow–peripheral blood dynamics in the disease state based on current emerging paradigms, part I. Math Biosci. 2016;274:83–93. https://doi.org/10.1016/j.mbs.2016.01.010.
Afenya EK, Ouifki R, Mundle SD. Mathematical modeling of bone marrow - peripheral blood dynamics in the disease state based on current emerging paradigms, part II. J Theor Biol. 2019;460:37–55. https://doi.org/10.1016/j.jtbi.2018.10.008.ThisarticleincorporatesCSCastheirowncompartmentinacompartmentalmodelforhematologicalcancers.
Elliott SL, Kose E, Lewis AL, Steinfeld AE, Zollinger EA. Modeling the stem cell hypothesis: investigating the effects of cancer stem cells and TGF-β on tumor growth. Math Biosci Eng. 2019;16(6):7177–94. https://doi.org/10.3934/mbe.2019360.
Gentry SN, Jackson TL. A mathematical model of cancer stem cell driven tumor initiation: implications of niche size and loss of homeostatic regulatory mechanisms. PLoS One. 2013;8(8): e71128. https://doi.org/10.1371/journal.pone.0071128.
Hu GM, Lee CY, Chen YY, Pang NN, Tzeng WJ. Mathematical model of heterogeneous cancer growth with an autocrine signalling pathway. Cell Prolif. 2012;45(5):445–55. https://doi.org/10.1111/j.1365-2184.2012.00835.x.
• Sehl ME, Wicha MS. Modeling of interactions between cancer stem cells and their microenvironment: predicting clinical response. Methods Mol Biol. 2018;1711:333–49. https://doi.org/10.1007/978-1-4939-7493-1_16. This paper presents a stochastic model for breast cancer stem cell niche, and investigate the trajectories of different cell types.
Yan H, Romero-Lopez M, Frieboes HB, Hughes CCW, Lowengrub JS. Multiscale modeling of glioblastoma suggests that the partial disruption of vessel/cancer stem cell crosstalk can promote tumor regression without increasing invasiveness. IEEE Trans Biomed Eng. 2017;64(3):538–48. https://doi.org/10.1109/TBME.2016.2615566.
Chen C, Baumann WT, Clarke R, Tyson JJ. Modeling the estrogen receptor to growth factor receptor signaling switch in human breast cancer cells. FEBS Lett. 2013;587(20):3327–34. https://doi.org/10.1016/j.febslet.2013.08.022.
• Forouzannia F, Enderling H, Kohandel M. Mathematical modeling of the effects of tumor heterogeneity on the efficiency of radiation treatment schedule. Bull Math Biol. 2018;80(2):283–93. https://doi.org/10.1007/s11538-017-0371-5. The authors present a deterministic and a stochastic models to test the effects of tumor heterogeneity on the outcome of radiation therapy.
Javadi A, Keighobadi F, Nekoukar V, Ebrahimi M. Finite-set model predictive control of melanoma cancer treatment using signaling pathway inhibitor of cancer stem cell. IEEE/ACM Trans Comput Biol Bioinform. 2019;PP. https://doi.org/10.1109/tcbb.2019.2940658.
• Nazari F, Pearson AT, Nör JE, Jackson TL. A mathematical model for IL-6-mediated, stem cell driven tumor growth and targeted treatment. PLoS Comput Biol. 2018;14(1):e1005920. https://doi.org/10.1371/journal.pcbi.1005920. In this article, the authors design a cellular and molecular differential equations model for IL-6 mediated stem cell growth and tumor initiation.
Sehl ME, Shimada M, Landeros A, Lange K, Wicha MS. Modeling of cancer stem cell state transitions predicts therapeutic response. PLoS One. 2015;10(9): e0135797. https://doi.org/10.1371/journal.pone.0135797.
• Sigal D, Przedborski M, Sivaloganathan D, Kohandel M. Mathematical modelling of cancer stem cell-targeted immunotherapy. Math Biosci. 2019;318:108269. https://doi.org/10.1016/j.mbs.2019.108269. The article incorporates differentiation and dedifferentiation of dendritic cells and T-cells which are trained to either attack the CSC or non-stem cancer cells.
Ward Rashidi MR, Mehta P, Bregenzer M, Raghavan S, Fleck EM, Horst EN, et al. Engineered 3D model of cancer stem cell enrichment and chemoresistance. Neoplasia. 2019;21(8):822–36. https://doi.org/10.1016/j.neo.2019.06.005.Thisarticleusesatwo-compartmentdifferentialequationsmodeltoanswerquestionsofchemoresistanceinovariancancer.
Werner B, Scott JG, Sottoriva A, Anderson AR, Traulsen A, Altrock PM. The cancer stem cell fraction in hierarchically organized tumors can be estimated using mathematical modeling and patient-specific treatment trajectories. Cancer Res. 2016;76(7):1705–13. https://doi.org/10.1158/0008-5472.Can-15-2069.
Eun K, Ham SW, Kim H. Cancer stem cell heterogeneity: origin and new perspectives on CSC targeting. BMB Rep. 2017;50(3):117–25. https://doi.org/10.5483/bmbrep.2017.50.3.222.
Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16(3):225–38. https://doi.org/10.1016/j.stem.2015.02.015.
Mukherjee S, Kong J, Brat DJ. Cancer stem cell division: when the rules of asymmetry are broken. Stem Cells Dev. 2015;24(4):405–16. https://doi.org/10.1089/scd.2014.0442.
Bu P, Chen K-Y, Chen Joyce H, Wang L, Walters J, Shin Yong J, et al. A microRNA miR-34a-regulated bimodal switch targets notch in colon cancer stem cells. Cell Stem Cell. 2013;12(5):602–15. https://doi.org/10.1016/j.stem.2013.03.002.
Enderling H. Cancer stem cells: small subpopulation or evolving fraction? Integr Biol (Camb). 2015;7(1):14–23. https://doi.org/10.1039/c4ib00191e.
Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–34. https://doi.org/10.1038/nm.4409.
López-Lázaro M. The stem cell division theory of cancer. Crit Rev Oncolo Hematol. 2018;123:95–113. https://doi.org/10.1016/j.critrevonc.2018.01.010.
Johnston MD, Edwards CM, Bodmer WF, Maini PK, Chapman SJ. Mathematical modeling of cell population dynamics in the colonic crypt and in colorectal cancer. Proc Natl Acad Sci. 2007;104(10):4008–13. https://doi.org/10.1073/pnas.0611179104.
Johnston MD, Edwards CM, Bodmer WF, Maini PK, Chapman SJ. Examples of mathematical modeling: tales from the crypt. Cell Cycle (Georgetown, Tex). 2007;6(17):2106–12. https://doi.org/10.4161/cc.6.17.4649.
• Renardy M, Jilkine A, Shahriyari L, Chou CS. Control of cell fraction and population recovery during tissue regeneration in stem cell lineages. J Theor Biol. 2018;445:33–50. https://doi.org/10.1016/j.jtbi.2018.02.017. This article presents a hierarchical stem cell lineage model and explores the necessity of feedback loops.
Benitez L, Barberis L, Vellon L, Condat CA. Understanding the influence of substrate when growing tumorspheres. BMC Cancer. 2021;21(1):276. https://doi.org/10.1186/s12885-021-07918-1.
Bessonov N, Pinna G, Minarsky A, Harel-Bellan A, Morozova N. Mathematical modeling reveals the factors involved in the phenomena of cancer stem cells stabilization. PLoS One. 2019;14(11): e0224787. https://doi.org/10.1371/journal.pone.0224787.ThisarticlepresentsaMarkovmodelforcelldivisionprobabilities.
Guo C, Ahmed S, Guo C, Liu X. Stability analysis of mathematical models for nonlinear growth kinetics of breast cancer stem cells. Math Methods Appl Sci. 2017;40(14):5332–48. https://doi.org/10.1002/mma.4389.
Scott JG, Dhawan A, Hjelmeland A, Lathia J, Chumakova A, Hitomi M, et al. Recasting the cancer stem cell hypothesis: unification using a continuum model of microenvironmental forces. Curr Stem Cell Rep. 2019;5(1):22–30. https://doi.org/10.1007/s40778-019-0153-0.
Cabrera MC, Hollingsworth RE, Hurt EM. Cancer stem cell plasticity and tumor hierarchy. World J Stem Cells. 2015;7(1):27–36. https://doi.org/10.4252/wjsc.v7.i1.27.
Carvalho J. Cell reversal from a differentiated to a stem-like state at cancer initiation. Front Oncol. 2020;10(541). https://doi.org/10.3389/fonc.2020.00541.
Nakano M, Kikushige Y, Miyawaki K, Kunisaki Y, Mizuno S, Takenaka K, et al. Dedifferentiation process driven by TGF-beta signaling enhances stem cell properties in human colorectal cancer. Oncogene. 2019;38(6):780–93. https://doi.org/10.1038/s41388-018-0480-0.
Wang P, Wan W-w, Xiong S-L, Feng H, Wu N. Cancer stem-like cells can be induced through dedifferentiation under hypoxic conditions in glioma, hepatoma and lung cancer. Cell Death Discov. 2017;3(1):16105. https://doi.org/10.1038/cddiscovery.2016.105.
• Zhou D, Luo Y, Dingli D, Traulsen A. The invasion of de-differentiating cancer cells into hierarchical tissues. PLoS Comput Biol. 2019;15(7):e1007167. https://doi.org/10.1371/journal.pcbi.1007167. This article presents a hierarchical model which allows for age dedifferentiation and argues that dedifferentiation plays a vital role in maintain populations of early age classes.
Jilkine A, Gutenkunst RN. Effect of dedifferentiation on time to mutation acquisition in stem cell-driven cancers. PLoS Comput Biol. 2014;10(3): e1003481. https://doi.org/10.1371/journal.pcbi.1003481.
Wang Y, Lo WC, Chou CS. Modelling stem cell ageing: a multi-compartment continuum approach. R Soc Open Sci. 2020;7(3): 191848. https://doi.org/10.1098/rsos.191848.
Deleyrolle LP, Ericksson G, Morrison BJ, Lopez JA, Burrage K, Burrage P, et al. Determination of somatic and cancer stem cell self-renewing symmetric division rate using sphere assays. PLoS One. 2011;6(1): e15844. https://doi.org/10.1371/journal.pone.0015844.
Lander AD, Gokoffski KK, Wan FYM, Nie Q, Calof AL. Cell lineages and the logic of proliferative control. PLoS Biol. 2009;7(1): e1000015. https://doi.org/10.1371/journal.pbio.1000015.
Rodriguez-Brenes IA, Komarova NL, Wodarz D. Evolutionary dynamics of feedback escape and the development of stem-cell-driven cancers. Proc Natl Acad Sci U S A. 2011;108(47):18983–8. https://doi.org/10.1073/pnas.1107621108.
• Weiss LD, van den Driessche P, Lowengrub JS, Wodarz D, Komarova NL. Effect of feedback regulation on stem cell fractions in tissues and tumors: Understanding chemoresistance in cancer. J Theor Biol. 2021;509:110499. https://doi.org/10.1016/j.jtbi.2020.110499. This article examines the impacts of feedback loops on the self-renewal, division, and death rates.
Lord BI, Gurney H, Chang J, Thatcher N, Crowther D, Dexter TM. Haemopoietic cell kinetics in humans treated with rGM-CSF. Int J Cancer. 1992;50(1):26–31. https://doi.org/10.1002/ijc.2910500107.
Afenya E, Mundle S. Hematologic disorders and bone marrow–peripheral blood dynamics. Math Model Nat Phenom. 2010;5(3):15–27.
• Benítez L, Barberis L, Condat CA. Modeling tumorspheres reveals cancer stem cell niche building and plasticity. Phys A Stat Mech Appl. 2019;533:121906. https://doi.org/10.1016/j.physa.2019.121906. This article examines the delicate balance between differentiated and cancer stem cells based on the probability of symmetric cell division.
Kaveh K, Kohandel M, Sivaloganathan S. Replicator dynamics of cancer stem cell: selection in the presence of differentiation and plasticity. Math Biosci. 2016;272:64–75. https://doi.org/10.1016/j.mbs.2015.11.012.
Konstorum A, Hillen T, Lowengrub J. Feedback regulation in a cancer stem cell model can cause an allee effect. Bull Math Biol. 2016;78(4):754–85. https://doi.org/10.1007/s11538-016-0161-5.
Mahdipour-Shirayeh A, Kaveh K, Kohandel M, Sivaloganathan S. Phenotypic heterogeneity in modeling cancer evolution. PLoS One. 2017;12(10): e0187000. https://doi.org/10.1371/journal.pone.0187000.
Wodarz D. Effect of cellular de-differentiation on the dynamics and evolution of tissue and tumor cells in mathematical models with feedback regulation. J Theor Biol. 2018;448:86–93. https://doi.org/10.1016/j.jtbi.2018.03.036.
Eastman B, Wodarz D, Kohandel M. The effects of phenotypic plasticity on the fixation probability of mutant cancer stem cells. J Theor Biol. 2020;503: 110384. https://doi.org/10.1016/j.jtbi.2020.110384.
Kharkar PS. Cancer Stem Cell (CSC) Inhibitors in oncology—a promise for a better therapeutic outcome: state of the art and future perspectives. J Med Chem. 2020;63(24):15279–307. https://doi.org/10.1021/acs.jmedchem.0c01336.
Lin M, Chang AE, Wicha M, Li Q, Huang S. Development and application of cancer stem cell-targeted vaccine in cancer immunotherapy. J Vaccines Vaccin. 2017;8(6):371. https://doi.org/10.4172/2157-7560.1000371.
• Brady R, Enderling H. Mathematical models of cancer: when to predict novel therapies, and when not to. Bull Math Biol. 2019;81(10):3722–31. https://doi.org/10.1007/s11538-019-00640-x. This article cautions about exploiting data in model development and stresses the importance of math and science collaboration.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no relevant conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Mathematical Models of Stem Cell Behaviour
Rights and permissions
About this article
Cite this article
Swanson, E.R., Elliott, S.L., Zollinger, E.A. et al. Cancer Stem Cell Division: Mathematical Models and Insights. Curr Stem Cell Rep 7, 204–211 (2021). https://doi.org/10.1007/s40778-021-00199-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40778-021-00199-1