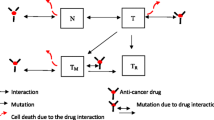
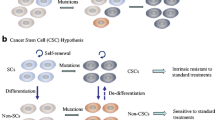
Abstract
Purpose of Review
Successful, durable cancer treatment is limited by drug resistance. Cancer stem cells (CSC) comprise (typically) a rare tumor subpopulation that contributes both intrinsic drug resistance and tumor re-initiation after therapy. Emerging evidence suggests that drug resistance is more complex than this single-cell level might suggest, and is likely governed by dynamics encompassing the entire tumor population. Here, we discuss the complexity of drug resistance by focusing on efforts that interface biology (wet lab) with mathematical modeling and simulation (dry lab) to study and explain the role of CSC and other cancer cells in the context of the entire ecosystem.
Recent Findings
Starting from biological evidence, we review the current state of cancer research from the perspective of the single-cell level, “The cancer cell,” its intrinsic physiopathology and its response to drug exposure. We discuss insufficiencies of this level of observation, in particular, the unaccounted for resistance to targeted therapies, and show why it is necessary to consider the entirety of the cell population, which is the only way to capture the role of biological heterogeneity. Importantly, we review how mathematical models have been implemented to elucidate mechanisms of drug resistance, and efforts made to validate biological experiments. Finally, we present emerging biological models, and therapeutic strategies inspired by mathematics, with the goal of improving the clinical management of cancer.
Summary
Over the past century, we have learned that cancer drug resistance is extraordinarily complex and requires an interdisciplinary scientific effort to unmask. The network of communication between and among cells within the diverse tumor heterogeneity drives acquired and intrinsic mechanisms of resistance. Harnessing biology and math to simulate, study, and explain the mechanisms of resistance, by considering the whole tumor population, is providing new clues to overcome it.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58.
Okabe S, Tauchi T, Ohyashiki K. Characteristics of dasatinib- and imatinib-resistant chronic myelogenous leukemia cells. Clin Cancer Res. 2008;14(19):6181–6.
Gorman MF, et al. Outcome for children treated for relapsed or refractory acute myelogenous leukemia (rAML): a Therapeutic Advances in Childhood Leukemia (TACL) Consortium study. Pediatr Blood Cancer. 2010;55(3):421–9.
de Rooij JD, Zwaan CM, van den Heuvel-Eibrink M. Pediatric AML: from biology to clinical management. J Clin Med. 2015;4(1):127–49.
Marjanovic ND, Weinberg RA, Chaffer CL. Cell plasticity and heterogeneity in cancer. Clin Chem. 2013;59(1):168–79.
Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–37.
American Cancer Society. Early history of cancer. 2014. https://www.cancer.org/cancer/cancer-basics/history-of-cancer/what-is-cancer.html.
Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–8.
Christakis P. The birth of chemotherapy at Yale. Bicentennial lecture series: surgery grand round. Yale J Biol Med. 2011;84(2):169–72.
Luria SE, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28(6):491–511.
Skipper HE. The forty-year-old mutation theory of Luria and Delbruck and its pertinence to cancer chemotherapy. Adv Cancer Res. 1983;40:331–63.
Law LW. Origin of the resistance of leukaemic cells to folic acid antagonists. Nature. 1952;169(4302):628–9.
Goldie JH, Coldman AJ. A mathematic model for relating the drug sensitivity of tumors to their spontaneous mutation rate. Cancer Treat Rep. 1979;63(11–12):1727–33.
Riordan JR, Ling V. Purification of P-glycoprotein from plasma membrane vesicles of Chinese hamster ovary cell mutants with reduced colchicine permeability. J Biol Chem. 1979;254(24):12701–5.
Flintoff WF, et al. Overproduction of dihydrofolate reductase and gene amplification in methotrexate-resistant Chinese hamster ovary cells. Mol Cell Biol. 1982;2(3):275–85.
Gambacorti-Passerini C. Part I: milestones in personalised medicine—imatinib. Lancet Oncol. 2008;9(6):600.
Mahon FX, et al. Selection and characterization of BCR-ABL positive cell lines with differential sensitivity to the tyrosine kinase inhibitor STI571: diverse mechanisms of resistance. Blood. 2000;96(3):1070–9.
Berrieman HK, Lind MJ, Cawkwell L. Do beta-tubulin mutations have a role in resistance to chemotherapy? Lancet Oncol. 2004;5(3):158–64.
Talpaz M, et al. Imatinib induces durable hematologic and cytogenetic responses in patients with accelerated phase chronic myeloid leukemia: results of a phase 2 study. Blood. 2002;99(6):1928–37.
Hirschmann-Jax C, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A. 2004;101(39):14228–33.
Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138(4):645–59.
Wan X, et al. Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene. 2007;26(13):1932–40.
• Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. A milestone paper that elicits transient, epigenetically controlled, drug resistance.
Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–44.
Goldman A, et al. Temporally sequenced anticancer drugs overcome adaptive resistance by targeting a vulnerable chemotherapy-induced phenotypic transition. Nat Commun. 2015;6:6139.
Pisco AO, et al. Non-Darwinian dynamics in therapy-induced cancer drug resistance. Nat Commun. 2013;4:2467.
O'Brien CA, Kreso A, Jamieson CH. Cancer stem cells and self-renewal. Clin Cancer Res: Off J Am Assoc Cancer Res. 2010;16(12):3113–20.
Patel P, Chen EI. Cancer stem cells, tumor dormancy, and metastasis. Front Endocrinol (Lausanne). 2012;3:125.
Trumpp A, Wiestler OD. Mechanisms of disease: cancer stem cells—targeting the evil twin. Nat Clin Pract Oncol. 2008;5(6):337–47.
Kern SE, Shibata D. The fuzzy math of solid tumor stem cells: a perspective. Cancer Res. 2007;67(19):8985–8.
Klonisch T, et al. Cancer stem cell markers in common cancers—therapeutic implications. Trends Mol Med. 2008;14(10):450–60.
Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–84.
Wichmann HE, Loeffler M. Mathematical modeling of cell proliferation: stem cell regulation in hemopoiesis. Vol. 1. Boca Raton: CRC Press; 1985.
Ganguly R, Puri IK. Mathematical model for the cancer stem cell hypothesis. Cell Prolif. 2006;39(1):3–14.
Turner C, et al. Characterization of brain cancer stem cells: a mathematical approach. Cell Prolif. 2009;42(4):529–40.
Michor F. Mathematical models of cancer stem cells. J Clin Oncol. 2008;26(17):2854–61.
Boman BM, et al. Symmetric division of cancer stem cells—a key mechanism in tumor growth that should be targeted in future therapeutic approaches. Clin Pharmacol Ther. 2007;81(6):893–8.
van Leeuwen IM, et al. Crypt dynamics and colorectal cancer: advances in mathematical modelling. Cell Prolif. 2006;39(3):157–81.
Merrell AJ, Stanger BZ. Adult cell plasticity in vivo: de-differentiation and transdifferentiation are back in style. Nat Rev Mol Cell Biol. 2016;17(7):413–25.
Heerboth S, et al. Use of epigenetic drugs in disease: an overview. Genet Epigenet. 2014;6:9–19.
Turner C, Kohandel M. Investigating the link between epithelial-mesenchymal transition and the cancer stem cell phenotype: a mathematical approach. J Theor Biol. 2010;265(3):329–35.
Chaffer CL, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108(19):7950–5.
Jilkine A, Gutenkunst RN. Effect of dedifferentiation on time to mutation acquisition in stem cell-driven cancers. PLoS Comput Biol. 2014;10(3):e1003481.
Kaveh K, Kohandel M, Sivaloganathan S. Replicator dynamics of cancer stem cell: selection in the presence of differentiation and plasticity. Math Biosci. 2016;272:64–75.
Goldman A. Tailoring combinatorial cancer therapies to target the origins of adaptive resistance. Mol Cell Oncol. 2016;3(1):e1030534.
Goldman, A., et al. Rationally designed 2-in-1 nanoparticles can overcome adaptive resistance in cancer. ACS Nano. 2016;10(6):5823–34.
Sun X, Bao J, Shao Y. Mathematical modeling of therapy-induced cancer drug resistance: connecting cancer mechanisms to population survival rates. Sci Rep. 2016;6:22498.
Tovar JD. Supramolecular construction of optoelectronic biomaterials. Acc Chem Res. 2013;46(7):1527–37.
Marusyk A, et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514(7520):54–8.
Moore LR, Rocap G, Chisholm SW. Physiology and molecular phylogeny of coexisting prochlorococcus ecotypes. Nature. 1998;393(6684):464–7.
Valaskova V, et al. Phylogenetic composition and properties of bacteria coexisting with the fungus Hypholoma fasciculare in decaying wood. ISME J. 2009;3(10):1218–21.
Hardin G. The competitive exclusion principle. Science. 1960;131(3409):1292–7.
Gerlinger M, Norton L, Swanton C. Acquired resistance to crizotinib from a mutation in CD74-ROS1. N Engl J Med. 2013;369(12):1172–3.
Zanoni M, et al. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci Rep. 2016;6:19103.
Nyga A, et al. A novel tissue engineered three-dimensional in vitro colorectal cancer model. Acta Biomater. 2013;9(8):7917–26.
Arai K, et al. A novel high-throughput 3D screening system for EMT inhibitors: a pilot screening discovered the EMT inhibitory activity of CDK2 inhibitor SU9516. PLoS One. 2016;11(9):e0162394.
Majumder B, et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat Commun. 2015;6:6169.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Aaron Goldman, Mohammad Kohandel, and Jean Clairambault declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Mathematical Models of Stem Cell Behavior
Rights and permissions
About this article
Cite this article
Goldman, A., Kohandel, M. & Clairambault, J. Integrating Biological and Mathematical Models to Explain and Overcome Drug Resistance in Cancer. Part 1: Biological Facts and Studies in Drug Resistance. Curr Stem Cell Rep 3, 253–259 (2017). https://doi.org/10.1007/s40778-017-0097-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40778-017-0097-1