Abstract
Background
Circular RNAs (circRNAs) are non-coding RNAs that have essential regulatory roles in the development of various tumors. This study explored whether circRNAs are involved in the progression of papillary thyroid carcinoma (PTC).
Methods
Differentially expressed circRNAs (DECs) in four pairs of PTC and matched normal thyroid tissues were screened using a circRNA microarray. The potential functions of dysregulated circRNAs were predicted by bioinformatic analyses. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was used to determine hsa_circ_0082003 expression in 80 pairs of PTC and matched normal thyroid tissues. Cell counting kit-8, colony formation, wound healing, and Transwell assays were performed to evaluate the biological functions of hsa_circ_0082003 in PTC cells. The role of hsa_circ_0082003 in PTC tumorigenesis in vivo was validated in nude mice.
Results
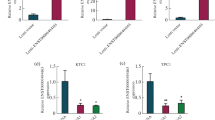
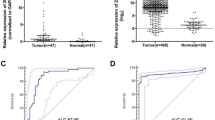
In total, 3150 DECs (2317 upregulated and 833 downregulated) were identified. Pathway enrichment analyses indicated that the dysregulated circRNAs may play roles in PTC development. RT-qPCR validation demonstrated that hsa_circ_0082003 expression was significantly increased in PTC tissues and correlated with poor clinicopathological parameters. Receiver operating characteristic curve analysis showed that hsa_circ_0082003 had good performance for diagnosing PTC and judging whether it was accompanied by lymph node metastasis. Knockdown of hsa_circ_0082003 inhibited PTC cell proliferation, migration, and invasion. Tumor formation assays in vivo showed that downregulation of hsa_circ_0082003 significantly suppressed the growth of PTC.
Conclusion
Hsa_circ_0082003 may serve as a novel diagnostic biomarker and potential therapeutic target for PTC.






Similar content being viewed by others
Data availability
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Abbreviations
- AUC:
-
Area under the receiver operating characteristic curve
- BP:
-
Biological process
- CC:
-
Cellular component
- CI:
-
Confidence interval
- circRNA:
-
Circular RNA
- DEC:
-
Differentially expressed circRNA
- ECM:
-
Extracellular matrix
- ETE:
-
Extra-thyroidal extension
- FBS:
-
Fetal bovine serum
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- LNM:
-
Lymph node metastasis
- MF:
-
Molecular function
- miRNA:
-
MicroRNA
- NC:
-
Negative control
- NS:
-
Not statistically significant
- PBS:
-
Phosphate buffered saline
- PTC:
-
Papillary thyroid carcinoma
- pTNM:
-
Pathological tumor, node, and metastasis
- ROC:
-
Receiver operating characteristic
- RPMI:
-
Roswell park memorial institute
- RT-qPCR:
-
Reverse transcription quantitative polymerase chain reaction
- shRNA:
-
Short hairpin RNA
- siRNA:
-
Small interfering RNA
- TC:
-
Thyroid carcinoma
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2022) Cancer statistics, 2022. CA Cancer J Clin 72:7–33. https://doi.org/10.3322/caac.21708
Haugen BR, Alexander EK, Bible KC, Doherty GM, Mandel SJ, Nikiforov YE, Pacini F, Randolph GW, Sawka AM, Schlumberger M, Schuff KG, Sherman SI, Sosa JA, Steward DL, Tuttle RM, Wartofsky L (2016) 2015 American Thyroid Association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: the American Thyroid Association guidelines task force on thyroid nodules and differentiated thyroid cancer. Thyroid 26:1–133. https://doi.org/10.1089/thy.2015.0020
Rui ZY, Liu Y, Zheng W, Wang X, Meng ZW, Tan J, Li N, Jia Q (2021) A retrospective study of the risk factors and the prognosis in patients with papillary thyroid carcinoma depending on the number of lymph node metastasis. Clin Exp Med 21:277–286. https://doi.org/10.1007/s10238-020-00675-8
Guang Y, He W, Zhang W, Zhang H, Zhang Y, Wan F (2021) Clinical study of ultrasonographic risk factors for central lymph node metastasis of papillary thyroid carcinoma. Front Endocrinol (Lausanne) 12:791970. https://doi.org/10.3389/fendo.2021.791970
Zhao H, Li H (2019) Meta-analysis of ultrasound for cervical lymph nodes in papillary thyroid cancer: diagnosis of central and lateral compartment nodal metastases. Eur J Radiol 112:14–21. https://doi.org/10.1016/j.ejrad.2019.01.006
Hsu MT, Coca-Prados M (1979) Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280:339–340. https://doi.org/10.1038/280339a0
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20:675–691. https://doi.org/10.1038/s41576-019-0158-7
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH (2020) Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer 19:172. https://doi.org/10.1186/s12943-020-01286-3
Li J, Sun D, Pu W, Wang J, Peng Y (2020) Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer 6:319–336. https://doi.org/10.1016/j.trecan.2020.01.012
Qiu L, Xu H, Ji M, Shang D, Lu Z, Wu Y, Tu Z, Liu H (2019) Circular RNAs in hepatocellular carcinoma: Biomarkers, functions and mechanisms. Life Sci 231:116660. https://doi.org/10.1016/j.lfs.2019.116660
Nie H, Wang Y, Liao Z, Zhou J, Ou C (2020) The function and mechanism of circular RNAs in gastrointestinal tumours. Cell Prolif 53:e12815. https://doi.org/10.1111/cpr.12815
Pei X, Chen SW, Long X, Zhu SQ, Qiu BQ, Lin K, Lu F, Xu JJ, Zhang PF, Wu YB (2020) circMET promotes NSCLC cell proliferation, metastasis, and immune evasion by regulating the miR-145–5p/CXCL3 axis. Aging (Albany, NY) 12:13038–13058. https://doi.org/10.18632/aging.103392
Gene Ontology Consortium (2021) The gene ontology resource: enriching a gold mine. Nucleic Acids Res 49:D325–D334. https://doi.org/10.1093/nar/gkaa1113
Geistlinger L, Csaba G, Santarelli M, Ramos M, Schiffer L, Turaga N, Law C, Davis S, Carey V, Morgan M, Zimmer R, Waldron L (2021) Toward a gold standard for benchmarking gene set enrichment analysis. Brief Bioinform 22:545–556. https://doi.org/10.1093/bib/bbz158
Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 73:3852–3856. https://doi.org/10.1073/pnas.73.11.3852
Patop IL, Wüst S, Kadener S (2019) Past, present, and future of circRNAs. EMBO J 38:e100836. https://doi.org/10.15252/embj.2018100836
Lei B, Tian Z, Fan W, Ni B (2019) Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci 16:292–301. https://doi.org/10.7150/ijms.28047
Chen L, Wang C, Sun H, Wang J, Liang Y, Wang Y, Wong G (2021) The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform 22:1706–1728. https://doi.org/10.1093/bib/bbaa001
Li S, Teng S, Xu J, Su G, Zhang Y, Zhao J, Zhang S, Wang H, Qin W, Lu ZJ, Guo Y, Zhu Q, Wang D (2019) Microarray is an efficient tool for circRNA profiling. Brief Bioinform 20:1420–1433. https://doi.org/10.1093/bib/bby006
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu Y, Dhanasekaran SM, Engelke CG, Cao X, Robinson DR, Nesvizhskii AI, Chinnaiyan AM (2019) The landscape of circular RNA in cancer. Cell 176:869-881.e13. https://doi.org/10.1016/j.cell.2018.12.021
Lei M, Zheng G, Ning Q, Zheng J, Dong D (2020) Translation and functional roles of circular RNAs in human cancer. Mol Cancer 19:30. https://doi.org/10.1186/s12943-020-1135-7
Liu W, Zhao J, Jin M, Zhou M (2019) circRAPGEF5 contributes to papillary thyroid proliferation and metastatis by regulation miR-198/FGFR1. Mol Ther Nucleic Acids 14:609–616. https://doi.org/10.1016/j.omtn.2019.01.003
Pan Y, Xu T, Liu Y, Li W, Zhang W (2019) Upregulated circular RNA circ_0025033 promotes papillary thyroid cancer cell proliferation and invasion via sponging miR-1231 and miR-1304. Biochem Biophys Res Commun 510:334–338. https://doi.org/10.1016/j.bbrc.2019.01.108
Zang J, Lu D, Xu A (2020) The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res 98:87–97. https://doi.org/10.1002/jnr.24356
Chen LL (2020) The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 21:475–490. https://doi.org/10.1038/s41580-020-0243-y
Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen W, Yuan X, Yin W, Xu J, Chen K, He C, Wei L (2020) CircRNA inhibits DNA damage repair by interacting with host gene. Mol Cancer 19:128. https://doi.org/10.1186/s12943-020-01246-x
Tellman TV, Cruz LA, Grindel BJ, Farach-Carson MC (2021) Cleavage of the perlecan-semaphorin 3A-plexin A1-Neuropilin-1 (PSPN) complex by matrix metalloproteinase 7/matrilysin triggers prostate cancer cell dyscohesion and migration. Int J Mol Sci 22:3218. https://doi.org/10.3390/ijms22063218
Kalluri R, LeBleu VS (2020) The biology, function, and biomedical applications of exosomes. Science 367:eaau6977. https://doi.org/10.1126/science.aau6977
Carneiro BA, El-Deiry WS (2020) Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol 17:395–417. https://doi.org/10.1038/s41571-020-0341-y
Du WW, Yang W, Li X, Fang L, Wu N, Li F, Chen Y, He Q, Liu E, Yang Z, Awan FM, Liu M, Yang BB (2020) The circular RNA circSKA3 binds integrin beta1 to induce invadopodium formation enhancing breast cancer invasion. Mol Ther 28:1287–1298. https://doi.org/10.1016/j.ymthe.2020.03.002
Tewari D, Priya A, Bishayee A, Bishayee A (2022) Targeting transforming growth factor-beta signalling for cancer prevention and intervention: recent advances in developing small molecules of natural origin. Clin Transl Med 12:e795. https://doi.org/10.1002/ctm2.795
Nersisyan S, Novosad V, Engibaryan N, Ushkaryov Y, Nikulin S, Tonevitsky A (2021) ECM-receptor regulatory network and its prognostic role in colorectal cancer. Front Genet 12:782699. https://doi.org/10.3389/fgene.2021.782699
Wu S, Chen M, Huang J, Zhang F, Lv Z, Jia Y, Cui YZ, Sun LZ, Wang Y, Tang Y, Verhoeft KR, Li Y, Qin Y, Lin X, Guan XY, Lam KO (2021) ORAI2 promotes gastric cancer tumorigenicity and metastasis through PI3K/Akt signaling and MAPK-dependent focal adhesion disassembly. Cancer Res 81:986–1000. https://doi.org/10.1158/0008-5472.CAN-20-0049
Theocharis AD, Karamanos NK (2019) Proteoglycans remodeling in cancer: underlying molecular mechanisms. Matrix Biol 75–76:220–259. https://doi.org/10.1016/j.matbio.2017.10.008
Manzella L, Stella S, Pennisi MS, Tirrò E, Massimino M, Romano C, Puma A, Tavarelli M, Vigneri P (2017) New insights in thyroid cancer and p53 family proteins. Int J Mol Sci 18:1325. https://doi.org/10.3390/ijms18061325
Goncalves MD, Hopkins BD, Cantley LC (2018) Phosphatidylinositol 3-kinase, growth disorders, and cancer. N Engl J Med 379:2052–2062. https://doi.org/10.1056/NEJMra1704560
Recondo G, Che J, Jänne PA, Awad MM (2020) Targeting MET dysregulation in cancer. Cancer Discov 10:922–934. https://doi.org/10.1158/2159-8290.CD-19-1446
Liu H, Deng H, Zhao Y, Li C, Liang Y (2018) LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J Exp Clin Cancer Res 37:279. https://doi.org/10.1186/s13046-018-0950-9
Guo R, Luo J, Chang J, Rekhtman N, Arcila M, Drilon A (2020) MET-dependent solid tumours – molecular diagnosis and targeted therapy. Nat Rev Clin Oncol 17:569–587. https://doi.org/10.1038/s41571-020-0377-z
Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, Cai JB, Yang X, Fan J, Ke AW, Zhou J, Shi GM (2020) Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer 19:92. https://doi.org/10.1186/s12943-020-01213-6
Acknowledgements
We would like to thank Editage (www.editage.cn) for English language editing.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Conceptualization and experimental design: YJ and JY; Experimentation: JY, JWF, WXW, and GFQ; Data analysis and manuscript writing: JY and JWF; Contribution of reagents/materials/analysis tools: JH, LZH, FW, and SYL; All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
This manuscript has not been published nor submitted for publication elsewhere. All authors have contributed significantly, and agree with the content of the manuscript. The authors report no proprietary or commercial interest in any product mentioned or concept discussed in this article.
Ethical approval
This study was approved by the Ethical Committee of Changzhou First People’s Hospital and was performed according to the ethical standards laid down in the 1964 Declaration of Helsinki.
Informed consent
Informed consent was obtained from all study participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
40618_2022_1922_MOESM1_ESM.xls
Table S1 Detailed information on the 10 most significantly upregulated circRNAs in PTC tissues as compared with matched normal thyroid tissues (XLS 24 KB)
40618_2022_1922_MOESM2_ESM.tif
Fig. S1 hsa_circ_0082003 has no significant effect on the proliferation, migration, and invasion of the human normal thyroid cell line in vitro. A RT-qPCR analysis of the transfection efficiency in the Nthy-ori 3–1 cell line. Relative expression levels were normalized to GAPDH expression. B CCK-8 assay of Nthy-ori 3–1 cells transfected with sh-hsa_circ_0082003 or sh-NC at the indicated time points. Wound healing assay (C) and Transwell invasion assay (D) of Nthy-ori 3–1 cells transfected with sh-hsa_circ_0082003 or sh-NC, respectively. NS, not statistically significant. ***P < 0.001 (TIF 9138 KB)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ye, J., Feng, JW., Wu, WX. et al. Microarray profiling identifies hsa_circ_0082003 as a novel tumor promoter for papillary thyroid carcinoma. J Endocrinol Invest 46, 509–522 (2023). https://doi.org/10.1007/s40618-022-01922-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40618-022-01922-3