Abstract
Purpose of review
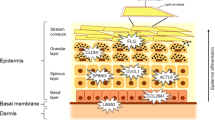
Mutations in the filaggrin gene can cause absent or reduced filaggrin protein, leading to impaired keratinization and skin barrier defect, which produce characteristic phenotypes. In this short review, we report current evidence on the topic with special reference to atopic dermatitis, suggest future directions, and discuss therapeutic implications.
Recent findings
Numerous candidate gene association studies, genome-wide association studies, studies on copy number variations, and, most recently, sequencing studies have confirmed the robust association of mutations in the filaggrin gene with atopic dermatitis, and have also linked these mutations with several other disorders.
Summary
Filaggrin gene defects remain the strongest identified genetic risk factors for atopic dermatitis. Taken in conjunction with other genes found to be associated with this condition, genetic screening and identification of individuals at risk for atopic dermatitis could lead to personalized therapy. Manipulation of genetic regulatory elements to increase the amount of filaggrin protein in deficient individuals is an attractive treatment option for the future.
Similar content being viewed by others
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
•• Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38(3):337–42 Landmark study that demonstrated the association of FLG LOF mutations with ichthyosis vulgaris.
Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38(4):441–446. Landmark study that demonstrated the association of common FLG LOF mutations with eczema/AD.
• Rodríguez E, Baurecht H, Herberich E, Wagenpfeil S, Brown SJ, Cordell HJ, et al. Meta-analysis of filaggrin polymorphisms in eczema and asthma: robust risk factors in atopic disease. J Allergy Clin Immunol. 2009;123(6):1361–70.e7. https://doi.org/10.1016/j.jaci.2009.03.036 Meta-analysis that demonstrated the strong association of FLG LOF mutations with eczema/AD and asthma in the context of AD.
•• Bin L, Leung DY. Genetic and epigenetic studies of atopic dermatitis. Allergy Asthma Clin Immunol. 2016;12:52 Review that summarizes the role of different biological mechanisms in the pathogenesis of AD.
Sandilands A, Sutherland C, Irvine AD, McLean WH. Filaggrin in the frontline: role in skin barrier function and disease. J Cell Sci. 2009;122(Pt 9):1285–94. https://doi.org/10.1242/jcs.033969.
Drucker AM, Wang AR, Li WQ, Sevetson E, Block JK, Qureshi AA. The burden of atopic dermatitis: summary of a report for the National Eczema Association. J Investig Dermatol. 2017;137(1):26–30. https://doi.org/10.1016/j.jid.2016.07.012.
Spergel JM, Paller AS. Atopic dermatitis and the atopic march. J Allergy Clin Immunol. 2003;112(6 Suppl):S118–27.
Kapoor R, Menon C, Hoffstad O, Bilker W, Leclerc P, Margolis DJ. The prevalence of atopic triad in children with physician-confirmed atopic dermatitis. J Am Acad Dermatol. 2008;58(1):68–73.
Barnes KC. An update on the genetics of atopic dermatitis: scratching the surface in 2009. J Allergy Clin Immunol. 2010;125(1):16–29.e1-11; quiz 30-1. https://doi.org/10.1016/j.jaci.2009.11.008.
• Elias PM, Hatano Y, Williams ML. Basis for the barrier abnormality in atopic dermatitis: outside-inside-outside pathogenic mechanisms. J Allergy Clin Immunol. 2008;121(6):1337–43. https://doi.org/10.1016/j.jaci.2008.01.022 This article discusses the outside-inside hypothesis of AD pathogenesis.
•• Stemmler S, Hoffjan S. Trying to understand the genetics of atopic dermatitis. Mol Cell Probes. 2016;30(6):374–85. https://doi.org/10.1016/j.mcp.2016.10.004 Review that discusses the FLG gene, the EDC and GWAS and exome-sequencing studies performed in the context of AD.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. https://doi.org/10.1038/nature19057.
Gimalova GF, Karunas AS, Fedorova YY, Khusnutdinova EK. The study of filaggrin gene mutations and copy number variation in atopic dermatitis patients from Volga-Ural region of Russia. Gene. 2016;591(1):85–9. https://doi.org/10.1016/j.gene.2016.06.054.
• Margolis DJ, Mitra N, Gochnauer H, Wubbenhorst B, D’Andrea K, Kraya A, et al. Uncommon filaggrin variants are associated with persistent atopic dermatitis in African Americans. J Investig Dermatol. 2018;138(7):1501–6. https://doi.org/10.1016/j.jid.2018.01.029 Longitudinal study that used massively parallel sequencing to demonstrate that uncommon LOF variants in FLG are associated with AD and with more persistent AD in children of African ancestry.
Margolis DJ, Mitra N, Wubbenhorst B, D'Andrea K, Kraya AA, Hoffstad O, et al. Association of filaggrin loss-of-function variants with race in children with atopic dermatitis. JAMA Dermatol. 2019;31:1269. https://doi.org/10.1001/jamadermatol.2019.1946.
Margolis DJ, Mitra N, Wubbenhorst B, Nathanson KL. Filaggrin sequencing and bioinformatics tools. Arch Dermatol Res. 2019;312:155–8. https://doi.org/10.1007/s00403-019-01956-3.
Margolis DJ, Gupta J, Apter AJ, Ganguly T, Hoffstad O, Papadopoulos M, et al. Filaggrin-2 variation is associated with more persistent atopic dermatitis in African American subjects. J Allergy Clin Immunol. 2014;133(3):784–9. https://doi.org/10.1016/j.jaci.2013.09.015.
Pigors M, Common JEA, Wong XFCC, Malik S, Scott CA, Tabarra N, et al. Exome sequencing and rare variant analysis reveals multiple filaggrin mutations in Bangladeshi families with atopic eczema and additional risk genes. J Investig Dermatol. 2018;138(12):2674–7. https://doi.org/10.1016/j.jid.2018.05.013.
• Brown SJ, Kroboth K, Sandilands A, Campbell LE, Pohler E, Kezic S, et al. Intragenic copy number variation within filaggrin contributes to the risk of atopic dermatitis with a dose-dependent effect. J Investig Dermatol. 2012;132(1):98–104. https://doi.org/10.1038/jid.2011.342 This study demonstrated that CNVs within FLG are also significant risk factors for AD.
Fernandez K, Asad S, Taylan F, Wahlgren CF, Bilcha KD, Nordenskjöld M, et al. Intragenic copy number variation in the filaggrin gene in Ethiopian patients with atopic dermatitis. Pediatr Dermatol. 2017;34(3):e140–1. https://doi.org/10.1111/pde.13095.
Quiggle AM, Goodwin ZA, Marfatia TR, Kumar MG, Ciliberto H, Bayliss SJ, et al. Low filaggrin monomer repeats in African American pediatric patients with moderate to severe atopic dermatitis. JAMA Dermatol. 2015;151(5):557–9. https://doi.org/10.1001/jamadermatol.2014.4916.
Wan J, Mitra N, Hoffstad OJ, Margolis DJ. Influence of FLG mutations and TSLP polymorphisms on atopic dermatitis onset age. Ann Allergy Asthma Immunol. 2017;118(6):737–738.e1. https://doi.org/10.1016/j.anai.2017.04.003.
Ziyab AH, Hankinson J, Ewart S, Schauberger E, Kopec-Harding K, Zhang H, et al. Epistasis between FLG and IL4R genes on the risk of allergic sensitization: results from two population-based birth cohort studies. Sci Rep. 2018;8(1):3221. https://doi.org/10.1038/s41598-018-21459-x.
Bisgaard H, Simpson A, Palmer CN, Bønnelykke K, McLean I, Mukhopadhyay S. Gene-environment interaction in the onset of eczema in infancy: filaggrin loss-of-function mutations enhanced by neonatal cat exposure. PLoS Med. 2008;5(6):e131. https://doi.org/10.1371/journal.pmed.0050131.
Schuttelaar ML, Kerkhof M, Jonkman MF, Koppelman GH, Brunekreef B, de Jongste JC, et al. Filaggrin mutations in the onset of eczema, sensitization, asthma, hay fever and the interaction with cat exposure. Allergy. 2009;64(12):1758–65. https://doi.org/10.1111/j.1398-9995.2009.02080.x.
Ait Bamai Y, Araki A, Nomura T, Kawai T, Tsuboi T, Kobayashi S, et al. Association of filaggrin gene mutations and childhood eczema and wheeze with phthalates and phosphorus flame retardants in house dust: the Hokkaido study on environment and children’s health. Environ Int. 2018;121(Pt 1):102–10. https://doi.org/10.1016/j.envint.2018.08.046.
Liljedahl ER, Wahlberg K, Lidén C, Albin M. Broberg K genetic variants of filaggrin are associated with occupational dermal exposure and blood DNA alterations in hairdressers. Sci Total Environ. 2019;653:45–54. https://doi.org/10.1016/j.scitotenv.2018.10.328.
Berg ND, Husemoen LL, Thuesen BH, Hersoug LG, Elberling J, Thyssen JP et al. Interaction between filaggrin null mutations and tobacco smoking in relation to asthma. J Allergy Clin Immunol. 2012;129(2):374–380.e1-2. doi: https://doi.org/10.1016/j.jaci.2011.08.045.
• Ziyab AH, Karmaus W, Holloway JW, Zhang H, Ewart S, Arshad SH. DNA methylation of the filaggrin gene adds to the risk of eczema associated with loss-of-function variants. J Eur Acad Dermatol Venereol. 2013;27(3):e420–3. https://doi.org/10.1111/jdv.12000 First study that identified the role of epigenetic mechanisms with regard to FLG and eczema/AD risk.
Esparza-Gordillo J, Matanovic A, Marenholz I, Bauerfeind A, Rohde K, Nemat K, et al. Maternal filaggrin mutations increase the risk of atopic dermatitis in children: an effect independent of mutation inheritance. PLoS Genet. 2015;11(3):e1005076. https://doi.org/10.1371/journal.pgen.1005076.
Elias PM, Williams ML. Basis for the gain and subsequent dilution of epidermal pigmentation during human evolution: the barrier and metabolic conservation hypotheses revisited. Am J Phys Anthropol. 2016;161(2):189–207. https://doi.org/10.1002/ajpa.23030.
• Thyssen JP, Bikle DD, Elias PM. Evidence that loss-of-function filaggrin gene mutations evolved in Northern Europeans to favor intracutaneous Vitamin D3 production. Evol Biol. 2014;41(3):388–96 Study that proposes that common LOF variants in FLG in Northern Europeans confer evolutionary advantage.
Eaaswarkhanth M, Xu D, Flanagan C, Rzhetskaya M, Hayes MG, Blekhman R, et al. Atopic dermatitis susceptibility variants in filaggrin Hitchhike hornerin selective sweep. Genome Biol Evol. 2016;8(10):3240–55.
Thyssen JP, Elias PM. It remains unknown whether filaggrin gene mutations evolved to increase cutaneous synthesis of vitamin D. Genome Biol Evol. 2017;9(4):900–1. https://doi.org/10.1093/gbe/evx049.
van den Oord RA, Sheikh A. Filaggrin gene defects and risk of developing allergic sensitisation and allergic disorders: systematic review and meta-analysis. BMJ. 2009;339:b2433. https://doi.org/10.1136/bmj.b2433.
Ponińska J, Samoliński B, Tomaszewska A, Raciborski F, Samel-Kowalik P, Walkiewicz A, et al. Filaggrin gene defects are independent risk factors for atopic asthma in a polish population: a study in ECAP cohort. PLoS One. 2011;6(2):e16933. https://doi.org/10.1371/journal.pone.0016933.
Weidinger S, O’Sullivan M, Illig T, Baurecht H, Depner M, Rodriguez E, et al. Filaggrin mutations, atopic eczema, hay fever, and asthma in children. J Allergy Clin Immunol. 2008;121(5):1203–1209.e1. https://doi.org/10.1016/j.jaci.2008.02.014.
Brown SJ, Asai Y, Cordell HJ, Campbell LE, Zhao Y, Liao H, et al. Loss-of-function variants in the filaggrin gene are a significant risk factor for peanut allergy. J Allergy Clin Immunol. 2011;127(3):661–7. https://doi.org/10.1016/j.jaci.2011.01.031.
Marenholz I, Grosche S, Kalb B, Rüschendorf F, Blümchen K, Schlags R et al Genome-wide association study identifies the SERPINB gene cluster as a susceptibility locus for food allergy. Nat Commun 2017;8(1):1056. doi: https://doi.org/10.1038/s41467-017-01220-0.
Bager P, Wohlfahrt J, Thyssen JP, Melbye M. Filaggrin genotype and skin diseases independent of atopic dermatitis in childhood. Pediatr Allergy Immunol. 2016;27(2):162–8. https://doi.org/10.1111/pai.12511.
Andersen YMF, Egeberg A, Balslev E, Jørgensen CLT, Szecsi PB, Stender S, et al. Filaggrin loss-of-function mutations, atopic dermatitis and risk of actinic keratosis: results from two cross-sectional studies. J Eur Acad Dermatol Venereol. 2017;31(6):1038–43. https://doi.org/10.1111/jdv.14172.
Handa S, Khullar G, Pal A, Kamboj P, De D. Filaggrin gene mutations in hand eczema patients in the Indian subcontinent: a prospective case-control study. Contact Dermatitis. 2019;80(6):359–64. https://doi.org/10.1111/cod.13233.
Kaae J, Szecsi PB, Meldgaard M, Espersen ML, Stender S, Johansen JD, et al. Individuals with complete filaggrin deficiency may have an increased risk of squamous cell carcinoma. Br J Dermatol. 2014;170(6):1380–1. https://doi.org/10.1111/bjd.12911.
Thyssen JP, Johansen JD, Carlsen BC, Linneberg A, Meldgaard M, Szecsi PB, et al. The filaggrin null genotypes R501X and 2282del4 seem not to be associated with psoriasis: results from general population study and meta-analysis. J Eur Acad Dermatol Venereol. 2012;26(6):782–4. https://doi.org/10.1111/j.1468-3083.2011.04107.x.
Hu Z, Xiong Z, Xu X, Li F, Lu L, Li W, et al. Loss-of-function mutations in filaggrin gene associate with psoriasis vulgaris in Chinese population. Hum Genet. 2012;131(7):1269–74. https://doi.org/10.1007/s00439-012-1155-5.
Chang YC, Wu WM, Chen CH, Hu CF, Hsu LA. Association between P478S polymorphism of the filaggrin gene and risk of psoriasis in a Chinese population in Taiwan. Arch Dermatol Res. 2008;300(3):133–7. https://doi.org/10.1007/s00403-007-0821-2.
Larsen KR, Johansen JD, Reibel J, Zachariae C, Rosing K, Pedersen AML. Filaggrin gene mutations and the distribution of filaggrin in oral mucosa of patients with oral lichen planus and healthy controls. J Eur Acad Dermatol Venereol. 2017;31(5):887–93. https://doi.org/10.1111/jdv.14098.
Thyssen JP, Andersen YMF, Balslev E, Szecsi PB, Stender S, Kaae J, et al. Loss-of-function mutations in filaggrin gene and malignant melanoma: a case-control study. J Eur Acad Dermatol Venereol. 2018;32(2):242–4. https://doi.org/10.1111/jdv.14532.
Kaae L, Thyssen JP, Johansen JD, Meldgaard M, Linneberg A, Allen M, et al. Filaggrin gene mutations and risk of basal cell carcinoma. Br J Dermatol. 2013;169(5):1162–4. https://doi.org/10.1111/bjd.12573.
Husemoen LL, Skaaby T, Jørgensen T, Thyssen JP, Meldgaard M, Szecsi PB, et al. No association between loss-of-function mutations in filaggrin and diabetes, cardiovascular disease, and all-cause mortality. PLoS One. 2013;8(12):e84293. https://doi.org/10.1371/journal.pone.0084293.
Joensen UN, Jørgensen N, Thyssen JP, Szecsi PB, Stender S, Petersen JH, et al. Urinary excretion of phenols, parabens and benzophenones in young men: associations to reproductive hormones and semen quality are modified by mutations in the filaggrin gene. Environ Int. 2018;121(Pt 1):365–74. https://doi.org/10.1016/j.envint.2018.09.020.
Kuang X, Sun L, Liu S, Zhao Z, Zhao D, Liu S, et al. Association of single nucleotide polymorphism rs2065955 of the filaggrin gene with susceptibility to Epstein-Barr virus-associated gastric carcinoma and EBV-negative gastric carcinoma. Virol Sin. 2016;31(4):306–13. https://doi.org/10.1007/s12250-016-3721-9.
Varbo A, Nordestgaard BG, Benn M. Filaggrin loss-of-function mutations as risk factors for ischemic stroke in the general population. J Thromb Haemost. 2017;15(4):624–35. https://doi.org/10.1111/jth.13644.
Yang Y, Liu W, Zhao Z, Zhang Y, Xiao H, Luo B. Filaggrin gene polymorphism associated with Epstein-Barr virus-associated tumors in China. Virus Genes. 2017;53(4):532–7. https://doi.org/10.1007/s11262-017-1463-x.
Margolis DJ, Apter AJ, Mitra N, Gupta J, Hoffstad O, Papadopoulos M, et al. Reliability and validity of genotyping filaggrin null mutations. J Dermatol Sci. 2013;70(1):67–8. https://doi.org/10.1016/j.jdermsci.2012.11.594.
Wong XFCC, Denil SLIJ, Foo JN, Chen H, Tay ASL, Haines RL, et al. Array-based sequencing of filaggrin gene for comprehensive detection of disease-associated variants. J Allergy Clin Immunol. 2018;141(2):814–6. https://doi.org/10.1016/j.jaci.2017.10.001.
Mócsai G, Gáspár K, Nagy G, Irinyi B, Kapitány A, Bíró T et al. Severe skin inflammation and filaggrin mutation similarly alter the skin barrier in patients with atopic dermatitis. Br J Dermatol. 2014;170(3):617–624. doi: https://doi.org/10.1111/bjd.12743. Study that demonstrated that skin barrier function in AD can be impaired due to severe skin inflammation independent of FLG mutation status.
Dajnoki Z, Béke G, Mócsai G, Kapitány A, Gáspár K, Hajdu K, et al. Immune-mediated skin inflammation is similar in severe atopic dermatitis patients with or without filaggrin mutation. Acta Derm Venereol. 2016;96(5):645–50. https://doi.org/10.2340/00015555-2272.
Hönzke S, Wallmeyer L, Ostrowski A, Radbruch M, Mundhenk L, Schäfer-Korting M, et al. Influence of Th2 cytokines on the cornified envelope, tight junction proteins, and ß-Defensins in filaggrin-deficient skin equivalents. J Investig Dermatol. 2016;136(3):631–9. https://doi.org/10.1016/j.jid.2015.11.007.
Guttman-Yassky E, Bissonnette R, Ungar B, Suárez-Fariñas M, Ardeleanu M, Esaki H, et al. Dupilumab progressively improves systemic and cutaneous abnormalities in patients with atopic dermatitis. J Allergy Clin Immunol. 2019 Jan;143(1):155–72. https://doi.org/10.1016/j.jaci.2018.08.022.
Tsuji G, Hashimoto-Hachiya A, Kiyomatsu-Oda M, Takemura M, Ohno F, Ito T, et al. Aryl hydrocarbon receptor activation restores filaggrin expression via OVOL1 in atopic dermatitis. Cell Death Dis. 2017 Jul 13;8(7):e2931. https://doi.org/10.1038/cddis.2017.322.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Jayanta Gupta reports being a co-investigator in a grant from NIAMS. David Margolis reports grants from NIAMS, personal fees from Pfizer, personal fees from Sanofi/Regeneron, grants from Valeant, and personal fees from Leo outside the submitted work.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Urticaria and Atopic Dermatitis
Rights and permissions
About this article
Cite this article
Gupta, J., Margolis, D.J. Filaggrin Gene Mutations with Special Reference to Atopic Dermatitis. Curr Treat Options Allergy 7, 403–413 (2020). https://doi.org/10.1007/s40521-020-00271-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40521-020-00271-x