Abstract
Introduction
Differentiation between intrapulmonary metastasis (IPM) and multiple primary lung cancers (MPLC) in patients with synchronous or metachronous lung tumor nodules is critical but challenging.
Objective
We proposed an algorithm to evaluate clonal origin based on trunk (initiating) versus branching drivers and the prevalence of mutations in lung adenocarcinomas.
Methods
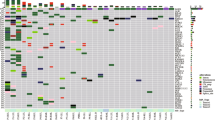
Driver mutations were examined using next-generation sequencing in five trunk driver genes (BRAF, EGFR, ERBB2, KRAS, and NRAS) and three branching driver genes (ATK1, PIK3CA, and TP53).
Results
Mutational profiling supported same clonality and likely same clonality, respectively, in 39 and 14 of 66 pairs of specimens with known identical clonal origin. Discordance of TP53 mutations (branching drivers) was observed in three pairs. Subsequent analyses of 30 pairs of synchronous or metachronous lung tumor nodules revealed different clonality and likely different clonality in 17 and 2 pairs, respectively, including three pairs with similar histomorphology; same clonality and likely same clonality in three and five pairs, respectively, including two pairs with different histomorphology; and inconclusive or noninformative results in three pairs.
Conclusion
While discordance of trunk drivers indicated MPLC in patients with synchronous or metachronous lung tumor nodules, discordance of branching drivers did not exclude IPM. Concordance of uncommon drivers supported IPM, whereas concordance of common drivers did not exclude MPLC. Additional recommendations from official organizations are needed to guide applications of molecular markers in defining clonality of multiple lung tumor nodules.


Similar content being viewed by others
References
Arai J, Tsuchiya T, Oikawa M, Mochinaga K, Hayashi T, Yoshiura K, et al. Clinical and molecular analysis of synchronous double lung cancers. Lung Cancer. 2012;77:281–7.
Loukeri AA, Kampolis CF, Ntokou A, Tsoukalas G, Syrigos K. Metachronous and synchronous primary lung cancers: diagnostic aspects, surgical treatment, and prognosis. Clin Lung Cancer. 2015;16:15–23.
Martini N, Melamed MR. Multiple primary lung cancers. J Thorac Cardiovasc Surg. 1975;70:606–12.
Shen KR, Meyers BF, Larner JM, Jones DR, American College of Chest Physicians.pecial treatment issues in lung cancer: ACCP evidence-based clinical practice guidelines (2nd edition). Chest. (2007);32(3 Suppl):290S–305S.
Detterbeck FC, Nicholson AG, Franklin WA, Marom EM, Travis WD, Girard N, et al. The IASLC lung cancer staging project: summary of proposals for revisions of the classification of lung cancers with multiple pulmonary sites of involvement in the forthcoming eighth edition of the TNM classification. J Thorac Oncol. 2016;11:639–50.
Detterbeck FC, Franklin WA, Nicholson AG, Girard N, Arenberg DA, Travis WD, et al. The IASLC lung cancer staging project: Background data and proposed criteria to distinguish separate primary lung cancers from metastatic foci in patients with two lung tumors in the forthcoming eighth edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11:651–65.
Detterbeck FC, Marom EM, Arenberg DA, Franklin WA, Nicholson AG, Travis WD, et al. The IASLC lung cancer staging project: Background data and proposals for the application of TNM staging rules to lung cancer presenting as multiple nodules with ground glass or lepidic features or a pneumonic type of involvement in the forthcoming eighth edition of the TNM classification. J Thorac Oncol. 2016;11:666–80.
Detterbeck FC, Bolejack V, Arenberg DA, Crowley J, Donington JS, Franklin WA, et al. The IASLC lung cancer staging project: Background data and proposals for the classification of lung cancer with separate tumor nodules in the forthcoming eighth edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11:681–92.
Rami-Porta R, Ball D, Crowley J, Giroux DJ, Jett J, Travis WD, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the T descriptors in the forthcoming (seventh) edition of the TNM classification for lung cancer. J Thorac Oncol. 2007;2:593–602.
Rami-Porta R, Asamura H, Travis WD, Rusch VW. In: Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK, et al., editors. AJCC Cancer Staging Manual. 8th ed. France: Springer International Publishing; 2017. p. 431–56.
Girard N, Deshpande C, Lau C, Finley D, Rusch V, Pao W, et al. Comprehensive histologic assessment helps to differentiate multiple lung primary nonsmall cell carcinomas from metastases. Am J Surg Pathol. 2009;33:1752–64.
Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 World Health Organization classification of lung tumors. Impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 2015;10:1243–60.
Lee HJ, Sung P, Yoen H, Kim S, Han S, et al. A novel algorithm to differentiate between multiple primary lung cancers and intrapulmonary metastasis in multiple lung cancers with multiple pulmonary sites of involvement. J Thorac Oncol. 2020;15:203–15.
Murphy SJ, Aubry MC, Harris FR, Halling GC, Johnson SH, Terra S, et al. Identification of independent primary tumors and intrapulmonary metastases using DNA rearrangements in non-small-cell lung cancer. J Clin Oncol. 2014;32:4050–8.
Wu C, Zhao C, Yang Y, He Y, Hou L, Li X, et al. High discrepancy of driver mutations in patients with NSCLC and synchronous multiple lung ground-glass nodules. J Thorac Oncol. 2015;10:778–83.
Schneider F, Derrick V, Davison JM, Strollo D, Incharoen P, Dacic S. Morphological and molecular approach to synchronous non-small cell lung carcinomas: impact on staging. Mod Pathol. 2016;29:735–42.
Liu Y, Zhang J, Li L, Yin G, Zhang J, Zheng S, et al. Genomic heterogeneity of multiple synchronous lung cancer. Nat Commun. 2016;7:13200.
Schneider F, Dacic S. Histopathologic and molecular approach to staging of multiple lung nodules. Transl Lung Cancer Res. 2017;6:540–9.
Asmar R, Sonett JR, Singh G, Mansukhani MM, Borczuk AC. Use of oncogenic driver mutations in staging of multiple primary lung carcinomas: a single-center experience. J Thorac Oncol. 2017;12:1524–35.
Patel SB, Kadi W, Walts AE, Marchevsky AM, Pao A, Aguiluz A, et al. Next-generation sequencing: a novel approach to distinguish multifocal primary lung adenocarcinomas from intrapulmonary metastases. J Mol Diagn. 2017;19:870–80.
Goto T, Hirotsu Y, Mochizuki H, Nakagomi T, Shikata D, Yokoyama Y, et al. Mutational analysis of multiple lung cancers: discrimination between primary and metastatic lung cancers by genomic profile. Oncotarget. 2017;8:31133–43.
Saab J, Zia H, Mathew S, Kluk M, Narula N, Fernandes H. Utility of genomic analysis in differentiating synchronous and metachronous lung adenocarcinomas from primary adenocarcinomas with intrapulmonary metastasis. Transl Oncol. 2017;10:442–9.
Roepman P, Ten Heuvel A, Scheidel KC, Sprong T, Heideman DAM, Seldenrijk KA, et al. Added Value of 50-gene panel sequencing to distinguish multiple primary lung cancers from pulmonary metastases: a systematic investigation. J Mol Diagn. 2018;20:436–45.
Murphy SJ, Harris FR, Kosari F, Barreto Siqueira Parrilha Terra S, Nasir A, Johnson SH, et al. Using genomics to differentiate multiple primaries from metastatic lung cancer. J Thorac Oncol. 2019;14:1567–82.
Chang JC, Alex D, Bott M, Tan KS, Seshan V, Golden A, et al. Comprehensive next-generation sequencing unambiguously distinguishes separate primary lung carcinomas from intra-pulmonary metastases: comparison with standard histopathologic approach. Clin Cancer Res. 2019;25:7113–25.
Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58.
Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–21.
Ahrendt SA, Decker PA, Alawi EA, Zhu Yr YR, Sanchez-Cespedes M, Yang SC, et al. Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer. 2001;92:1525–30.
Zhang YL, Yuan JQ, Wang KF, Fu XH, Han XR, Threapleton D, et al. The prevalence of EGFR mutation in patients with non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget. 2016;7:78985–93.
de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–6.
De Marchi F, Haley L, Fryer H, brahim J, Beierl K, Zheng G, et al. Clinical validation of coexisting activating mutations within EGFR, mitogen-activated protein kinase and phosphatidylinositol 3-kinase pathways in lung cancers. Arch Pathol Lab Med. 2019;143:174–82.
Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–31.
Illei PB, Belchis D, Tseng LH, Nguyen D, De Marchi F, Haley L, et al. Clinical mutational profiling of 1006 lung cancers by next generation sequencing. Oncotarget. 2017;8:96684–96.
Yao Z, Yaeger R, Rodrik-Outmezguine VS, Tao A, Torres NM, Chang MT, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature. 2017;548:234–8.
Lokhandwala PM, Tseng LH, Rodriguez E, Zheng G, Pallavajjalla A, Gocke CD, et al. Clinical mutational profiling and categorization of BRAF mutations in melanomas using next generation sequencing. BMC Cancer. 2019;19:665.
Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–67.
Nieto P, Ambrogio C, Esteban-Burgos L, Gómez-López G, Blasco MT, Yao Z, et al. A Braf kinase-inactive mutant induces lung adenocarcinoma. Nature. 2017;548(7666):239–43.
Tseng LH, De Marchi F, Pallavajjalla A, Rodriguez E, Xian R, Belchis D, et al. Clinical validation of discordant trunk driver mutations in paired primary and metastatic lung cancer specimens. Am J Clin Pathol. 2019;152:570–81.
Zheng G, Lin MT, Lokhandwala PM, Beierl K, Netto GJ, Gocke CD, et al. Clinical mutational profiling of bone metastases of lung and colon carcinoma and malignant melanoma using next-generation sequencing. Cancer Cytopathol. 2016;124:744–53.
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6.
Lin MT, Mosier SL, Thiess M, Beierl KF, Debeljak M, Tseng LH, et al. Clinical validation of KRAS, BRAF, and EGFR mutation detection using next-generation sequencing. Am J Clin Pathol. 2014;141:856–66.
Sikkink SK, Liloglou T, Maloney P, Gosney JR, Field JK. In-depth analysis of molecular alterations within normal and tumour tissue from an entire bronchial tree. Int J Oncol. 2003;22:589–95.
Gazdar AF, Minna JD. Multifocal lung cancers-clonality vs field cancerization and does it matter? J Natl Cancer Inst. 2009;101:541–3.
Vignot S, Frampton GM, Soria JC, Commo F, Brambilla C, Palmer G, et al. Next-generation sequencing reveals high concordance of recurrent somatic alterations between primary tumor and metastases from patients with non-small-cell lung cancer. J Clin Oncol. 2013;31:2167–72.
Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011;29:2972–7.
Wang S, Wang Z. Meta-analysis of epidermal growth factor receptor and KRAS gene status between primary and corresponding metastatic tumours of non-small cell lung cancer. Clin Oncol (R Coll Radiol). 2015;27:30–9.
Pfeifer JD, Liu J. Rate of occult specimen provenance complications in routine clinical practice. Am J Clin Pathol. 2013;139:93–100.
Chang YL, Wu CT, Lin SC, Hsiao CF, Jou YS, Lee YC. Clonality and prognostic implications of p53 and epidermal growth factor receptor somatic aberrations in multiple primary lung cancers. Clin Cancer Res. 2007;13:52–8.
van Rens MT, Eijken EJ, Elbers JR, Lammers JW, Tilanus MG, Slootweg PJ. p53 mutation analysis for definite diagnosis of multiple primary lung carcinoma. Cancer. 2002;94:188–96.
Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13.
Takahashi Y, Shien K, Tomida S, Oda S, Matsubara T, Sato H, et al. Comparative mutational evaluation of multiple lung cancers by multiplex oncogene mutation analysis. Cancer Sci. 2018;109:3634–42.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
The authors have no funding to disclose.
Conflict of interest
Erika F. Rodriguez (EFR), Li-Hui Tseng (LHT), Federico De Marchi (FM), Jialing Haung (JH), Deborah Belchis (DB), Rena Xian (RX), Christopher D. Gocke (CDG), James R. Eshleman (JRE), Peter B. Illei (PBI), and Ming-Tseh Lin (MTL) have no conflicts of interest that are directly relevant to the content of this article.
Ethical approval and informed consent
This retrospective study for quality improvement was approved by the institutional review board on “Histopathological, immunohistochemical and molecular analysis of pulmonary tumors” (IRB Number 00063529) and conducted according to the principles of the Declaration of Helsinki.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rodriguez, E.F., Tseng, LH., De Marchi, F. et al. Clonal Origin Evaluated by Trunk and Branching Drivers and Prevalence of Mutations in Multiple Lung Tumor Nodules. Mol Diagn Ther 24, 461–472 (2020). https://doi.org/10.1007/s40291-020-00471-w
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-020-00471-w