Abstract
Background
Circulating free DNA in plasma is an alternative source of tumor-derived DNA that can be a surrogate for tissue epidermal growth factor receptor (EGFR) testing.
Objective
We evaluated the analytical performance of the cobas® EGFR Mutation Test v2 (cobas test), a real-time polymerase chain reaction assay designed to detect defined EGFR gene mutations in plasma from patients with advanced non-small cell lung cancer (NSCLC).
Methods
We used K2-ethylenediaminetetraacetic acid plasma samples from NSCLC patients and healthy donors (HDs), along with cell line DNA. Results from a complete technical performance evaluation are described, including a comparison between NSCLC and HD plasma to support the use of surrogate samples and an independent confirmation of the limit of detection (LoD).
Results
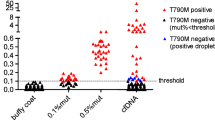
The cobas test reported an overall percent agreement of approximately 88% for plasma samples when compared with a next-generation sequencing method. The LoD for all EGFR mutations was ≤ 100 copies/mL for plasma samples. An external study confirmed the LoD for exon 19 deletion, L858R, and T790M at ≤ 100 copies/mL using samples derived from NSCLC patient specimens. The cobas test showed linearity between at least 50 and 10,000 copies/mL for plasma samples. An internal repeatability study reported a correct call accuracy of 99.2% for plasma samples. The performance of the cobas test is equivalent when using sheared or intact cell line DNA diluted into either HD plasma or NSCLC patient plasma.
Conclusions
The cobas test is a sensitive, robust, and accurate assay that delivers reproducible results.

Similar content being viewed by others
Availability of data and material
Associated datasets will not be deposited.
References
Union for International Cancer Control. New global cancer data: GLOBOCAN 2018. 2018. https://www.uicc.org/news/new-global-cancer-data-globocan-2018. Accessed 24 July 2019.
U.S. Cancer Statistics Working Group. U.S. Cancer Statistics Data Visualizations Tool, based on November 2018 submission data (1999-2016): U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute. 2018. https://gis.cdc.gov/Cancer/USCS/DataViz.html.
Novello S, Barlesi F, Califano R, Cufer T, Ekman S, Levra MG, et al. Metastatic non-small-cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27(suppl 5):v1–27. https://doi.org/10.1093/annonc/mdw326.
Hodoglugil U, Carrillo MW, Hebert JM, Karachaliou N, Rosell RC, Altman RB, et al. PharmGKB summary: very important pharmacogene information for the epidermal growth factor receptor. Pharmacogenet Genom. 2013;23(11):636–42. https://doi.org/10.1097/FPC.0b013e3283655091.
Morgensztern D, Politi K, Herbst RS. EGFR mutations in non-small-cell lung cancer: find, divide, and conquer. JAMA Oncol. 2015;1(2):146–8. https://doi.org/10.1001/jamaoncol.2014.278.
Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–500. https://doi.org/10.1126/science.1099314.
Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28(Suppl 1):S24–31. https://doi.org/10.1038/onc.2009.198.
Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39. https://doi.org/10.1056/NEJMoa040938.
Gray JE, Okamoto I, Sriuranpong V, Vansteenkiste J, Imamura F, Lee JS, et al. Tissue and plasma EGFR mutation analysis in the FLAURA trial: osimertinib versus comparator EGFR tyrosine kinase inhibitor as first-line treatment in patients with EGFR-mutated advanced non-small cell lung cancer. Clin Cancer Res. 2019;25(22):6644–52. https://doi.org/10.1158/1078-0432.Ccr-19-1126.
Jänne PA, Yang JC, Kim DW, Planchard D, Ohe Y, Ramalingam SS, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372(18):1689–99. https://doi.org/10.1056/NEJMoa1411817.
Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–57. https://doi.org/10.1056/NEJMoa0810699.
Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–46. https://doi.org/10.1016/s1470-2045(11)70393-x.
Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378(2):113–25. https://doi.org/10.1056/NEJMoa1713137.
Yang JC, Wu YL, Schuler M, Sebastian M, Popat S, Yamamoto N, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015;16(2):141–51. https://doi.org/10.1016/s1470-2045(14)71173-8.
Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. https://doi.org/10.1371/journal.pmed.0020073.
Goss G, Tsai C-M, Shepherd FA, Bazhenova L, Lee JS, Chang G-C, et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol. 2016;17(12):1643–52. https://doi.org/10.1016/S1470-2045(16)30508-3.
Xue C, Hu Z, Jiang W, Zhao Y, Xu F, Huang Y, et al. National survey of the medical treatment status for non-small cell lung cancer (NSCLC) in China. Lung Cancer. 2012;77(2):371–5. https://doi.org/10.1016/j.lungcan.2012.04.014.
Yoon HJ, Lee HY, Lee KS, Choi YL, Ahn MJ, Park K, et al. Repeat biopsy for mutational analysis of non-small cell lung cancers resistant to previous chemotherapy: adequacy and complications. Radiology. 2012;265(3):939–48. https://doi.org/10.1148/radiol.12112613.
Wiener RS, Wiener DC, Gould MK. Risks of transthoracic needle biopsy: how high? Clin Pulm Med. 2013;20(1):29–35. https://doi.org/10.1097/CPM.0b013e31827a30c1.
Davenport L. EGFR testing not done in 25% of lung cancer patients. Medscape Medical News; 2015. https://www.medscape.com/viewarticle/843332. Accessed 17 Jan 2020.
Malapelle U, Sirera R, Jantus-Lewintre E, Reclusa P, Calabuig-Farinas S, Blasco A, et al. Profile of the Roche cobas® EGFR mutation test v2 for non-small cell lung cancer. Expert Rev Mol Diagn. 2017;17(3):209–15. https://doi.org/10.1080/14737159.2017.1288568.
U.S. Food and Drug Administration. Premarket approval (PMA): cobas EGFR Mutation Test v2. 2019. https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P120019S016. Accessed 19 Sep 2019.
U.S. Food and Drug Administration. Premarket approval (PMA): cobas EGFR Mutation Test v2. 2016. https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P150047. Accessed 19 Sep 2019.
Jenkins S, Yang JC, Ramalingam SS, Yu K, Patel S, Weston S, et al. Plasma ctDNA analysis for detection of the EGFR T790M mutation in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2017;12(7):1061–70. https://doi.org/10.1016/j.jtho.2017.04.003.
Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376(7):629–40. https://doi.org/10.1056/NEJMoa1612674.
Wu YL, Lee V, Liam CK, Lu S, Park K, Srimuninnimit V, et al. Clinical utility of a blood-based EGFR mutation test in patients receiving first-line erlotinib therapy in the ENSURE, FASTACT-2, and ASPIRATION studies. Lung Cancer. 2018;126:1–8. https://doi.org/10.1016/j.lungcan.2018.10.004.
Inc Roche Molecular Systems. cobas® EGFR mutation test v2. Branchburg: Roche Molecular Systems Inc.; 2015.
Clinical and Laboratory Standards Institute. EP17-A2. Evaluation of detection capability for clinical laboratory measurement procedures; approved guideline - second edition. Wayne: Clinical and Laboratory Standards Institute; 2012.
Clinical and Laboratory Standards Institute. EP06-A. Evaluation of the linearity of quantitative measurement procedures: a statistical approach; approved guideline. Wayne: Clinical and Laboratory Standards Institute; 2003.
Clinical and Laboratory Standards Institute. EP07-A2. Interference testing in clinical chemistry; approved guideline—second edition. Wayne: Clinical and Laboratory Standards Institute; 2005.
Duan H, Lu J, Lu T, Gao J, Zhang J, Xu Y, et al. Comparison of EGFR mutation status between plasma and tumor tissue in non-small cell lung cancer using the Scorpion ARMS method and the possible prognostic significance of plasma EGFR mutation status. Int J Clin Exp Pathol. 2015;8(10):13136–45.
Wiener RS, Schwartz LM, Woloshin S, Welch HG. Population-based risk for complications after transthoracic needle lung biopsy of a pulmonary nodule: an analysis of discharge records. Ann Intern Med. 2011;155(3):137–44. https://doi.org/10.7326/0003-4819-155-3-201108020-00003.
Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. https://doi.org/10.1126/scitranslmed.3007094.
Szpechcinski A, Chorostowska-Wynimko J, Struniawski R, Kupis W, Rudzinski P, Langfort R, et al. Cell-free DNA levels in plasma of patients with non-small-cell lung cancer and inflammatory lung disease. Br J Cancer. 2015;113(3):476–83. https://doi.org/10.1038/bjc.2015.225.
Esposito A, Criscitiello C, Trapani D, Curigliano G. The emerging role of “liquid biopsies”, circulating tumor cells, and circulating cell-free tumor DNA in lung cancer diagnosis and identification of resistance mutations. Curr Oncol Rep. 2017;19(1):1. https://doi.org/10.1007/s11912-017-0564-y.
Heeke S, Benzaquen J, Hofman V, Ilie M, Allegra M, Long-Mira E, et al. Critical assessment in routine clinical practice of liquid biopsy for EGFR status testing in non-small-cell lung cancer: a single-laboratory experience (LPCE, Nice, France). Clin Lung Cancer. 2020;21(1):56–65.e8. https://doi.org/10.1016/j.cllc.2019.07.010.
Keppens C, Palma JF, Das PM, Scudder S, Wen W, Normanno N, et al. Detection of EGFR variants in plasma: a multilaboratory comparison of a real-time PCR EGFR mutation test in Europe. J Mol Diagn. 2018;20(4):483–94. https://doi.org/10.1016/j.jmoldx.2018.03.006.
Catalogue of Somatic Mutations in Cancer (COSMIC). v.51. 2011. https://cancer.sanger.ac.uk/cosmic. Accessed 19 Sep 2019.
Acknowledgements
We would like to acknowledge Sweta Shah, Johnny Shyu, Jingchuan Li, Robert Current, Taraneh Rehage, Misgana Bogale, and Yiqiao Wu (all of Roche Molecular Systems) for their contributions during the execution of this study. Funding for this study was provided by Roche Molecular Systems (Pleasanton, CA, USA). COBAS is a trademark of Roche. All other trademarks are the property of their respective owners.
Author information
Authors and Affiliations
Contributions
All authors contributed to the visualization, writing, review, and editing of the manuscript. PO was responsible for conceptualization, methodology, investigation, validation, formal analysis, and data curation. TM, KDM, and JF were responsible for methodology, validation, investigation, formal analysis, and data curation. HH was responsible for conceptualization, investigation, and data curation. WW was responsible for conceptualization, methodology, validation, investigation, and data curation. KY was responsible for conceptualization, validation, and data curation. SS was responsible for conceptualization. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Funding
This study was funded by Roche Molecular Solutions, Pleasanton, CA, USA.
Conflict of interest
Patrick O’Donnell is an employee of Roche Molecular Solutions. Theresa May is an employee of Roche Molecular Solutions. Kelli DeMartin is an employee of Roche Molecular Solutions. Jane Ferguson was previously an employee of Roche Molecular Systems until September 2018. Harkanwal Halait is an employee of Roche Molecular Solutions. Wei Wen is an employee of Roche Molecular Solutions and has previously received patent authorization for patents related to the cobas test. Karen Yu is an employee of Roche Molecular Solutions. Sid Scudder is an employee of Roche Molecular Solutions and owns stock in F. Hoffmann-La Roche AG.
Ethics approval
K2-EDTA plasma specimens from NSCLC patients and healthy donors were obtained from commercial vendors and collaborators with appropriate informed consent.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
O’Donnell, P., May, T., DeMartin, K. et al. Performance Characteristics of a Real-Time Polymerase Chain Reaction Assay for the Detection of Epidermal Growth Factor Receptor (EGFR) Mutations in Plasma Samples of Non-Small Cell Lung Cancer (NSCLC) Patients. Mol Diagn Ther 24, 451–460 (2020). https://doi.org/10.1007/s40291-020-00458-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-020-00458-7