Abstract
Background and Objective
Inherited long QT syndrome (LQTS) is a cardiac channelopathy associated with a high risk of sudden death. The prevalence has been estimated at close to 1:2,000. Due to large cohorts to investigate and high rate of private mutations, mutational screening must be performed using an extremely sensitive and specific detection method. Mutational screening is crucial as this may have implications for therapy and management of LQTS patients.
Methods
Next-generation sequencing (NGS) workflow based on a custom AmpliSeq™ panel was designed for sequencing the five most prevalent cardiomyopathy-causing genes (KCNQ1, KCNH2, SCN5A, KCNE1, KCNE2) on Ion PGM™ Sequencer. A cohort of 30 previously studied patients was screened to evaluate this strategy in terms of sensitivity, specificity, practicability, and cost. In silico analysis was performed using NextGENe® software.
Results
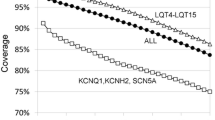
Our AmpliSeq™ custom panel allowed us to explore 86 % of targeted sequences efficiently. Using adjusted alignment settings, all genetic variants (40 substitutions, 17 indels) present in covered regions and previously detected by high-resolution melt (HRM)/sequencing were readily identified. Uncovered targeted regions, which were mainly located in KCNH2, were further analyzed by HRM/sequencing strategy. Complete molecular investigation was performed faster and cheaper than with previously used mutation detection methods.
Conclusion
Finally, these results suggested that our new NGS approach based on AmpliSeq™ libraries and Ion PGM™ sequencing is a highly efficient, fast, and cheap high-throughput mutation detection method that is ready to be deployed in clinical laboratories. This method will allow fast identification of LQTS mutations that will have further implications for therapeutics.

Similar content being viewed by others
References
Abriel H, Zaklyazminskaya EV. Cardiac channelopathies: genetic and molecular mechanisms. Gene. 2013;517:1–11.
Schwartz PJ, Stramba-Badiale M, Crotti L, et al. Prevalence of the congenital long-QT syndrome. Circulation. 2009;120:1761–7.
Ruan Y, Liu N, Napolitano C, Priori SG. Therapeutic strategies for long-QT syndrome: does the molecular substrate matter? Circ Arrhythm Electrophysiol. 2008;1:290–7.
Millat G, Chanavat V, Créhalet H, Rousson R. Development of a high resolution melting method for the detection of genetic variations in Long QT Syndrome. Clin Chim Acta. 2011;412:203–7.
Millat G, Chevalier P, Restier-Miron L, et al. Spectrum of pathogenic mutations and associated polymorphisms in a cohort of 44 unrelated patients with long QT syndrome. Clin Genet. 2006;70:214–27.
Millat G, Chanavat V, Rodriguez-Lafrasse C, Rousson R. Rapid, sensitive and inexpensive detection of SCN5A genetic variations by high resolution melting analysis. Clin Biochem. 2009;42:491–9.
Sikkema-Raddatz B, Johansson LF, de Boer EN, et al. Targeted next-generation sequencing can replace Sanger sequencing in clinical diagnostics. Hum Mutat. 2013;34:1035–42.
Ware JS, John S, Roberts AM, et al. Next generation diagnostics in inherited arrhythmia syndromes: a comparison of two approaches. J Cardiovasc Transl Res. 2013;6:94–103.
Lieve KV, Williams L, Daly A, et al. Results of genetic testing in 855 consecutive unrelated patients referred for long QT syndrome in a clinical laboratory. Genet Test Mol Biomarkers. 2013;17:553–61.
Tarabeux J, Zeitouni B, Moncoutier V, et al. Streamlined ion torrent PGM-based diagnostics: BRCA1 and BRCA2 genes as a model. Eur J Hum Genet. 2014;22:535–41.
AbouTayoun AN, Tunkey CD, Pugh TJ, et al. A comprehensive assay for CFTR mutational analysis using next-generation sequencing. Clin Chem. 2013;59:1481–8.
Elliott AM, Radecki J, Moghis B, Li X, Kammesheidt A. Rapid detection of the ACMG/ACOG-recommended 23 CFTR disease-causing mutations using ion torrent semiconductor sequencing. J Biomol Tech. 2012;23:24–30.
Yeo ZX, Chan M, Yap YS, Ang P, Rozen S, Lee AS. Improving indel detection specificity of the Ion Torrent PGM benchtop sequencer. PLoS One. 2012;7:e45798.
Chin EL, da Silva C, Hegde M. Assessment of clinical analytical sensitivity and specificity of next-generation sequencing for detection of simple and complex mutations. BMC Genet. 2013;14:6.
Barc J, Briec F, Schmitt S, et al. Screening for copy number variation in genes associated with the long QT syndrome: clinical relevance. J Am Coll Cardiol. 2011;57:40–7.
Acknowledgments
This work was supported by the French Ministry of Research (Diagnosis Network on Neuromuscular Diseases). The authors have no conflicts of interest to declare that are directly relevant to the content of this article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Millat, G., Chanavat, V. & Rousson, R. Evaluation of a New High-Throughput Next-Generation Sequencing Method Based on a Custom AmpliSeq™ Library and Ion Torrent PGM™ Sequencing for the Rapid Detection of Genetic Variations in Long QT Syndrome. Mol Diagn Ther 18, 533–539 (2014). https://doi.org/10.1007/s40291-014-0099-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-014-0099-y