Abstract
Introduction
Outcomes in type 2 diabetes mellitus (T2DM) could be optimized by identifying which treatments are likely to produce the greatest improvements in glycemic control for each patient.
Objectives
We aimed to identify patient characteristics associated with achieving and maintaining a target glycated hemoglobin (HbA1c) of ≤ 7% using machine learning methodology to analyze clinical trial data on combination therapy for T2DM. By applying a new machine learning methodology to an existing clinical dataset, the practical application of this approach was evaluated and the potential utility of this new approach to clinical decision making was assessed.
Methods
Data were pooled from two phase III, randomized, double-blind, parallel-group studies of empagliflozin/linagliptin single-pill combination therapy versus each monotherapy in patients who were treatment-naïve or receiving background metformin. Descriptive analysis was used to assess univariate associations between HbA1c target categories and each baseline characteristic. After the descriptive analysis results, a machine learning analysis was performed (classification tree and random forest methods) to estimate and predict target categories based on patient characteristics at baseline, without a priori selection.
Results
In the descriptive analysis, lower mean baseline HbA1c and fasting plasma glucose (FPG) were both associated with achieving and maintaining the HbA1c target. The machine learning analysis also identified HbA1c and FPG as the strongest predictors of attaining glycemic control. In contrast, covariates including body weight, waist circumference, blood pressure, or other variables did not contribute to the outcome.
Conclusions
Using both traditional and novel data analysis methodologies, this study identified baseline glycemic status as the strongest predictor of target glycemic control attainment. Machine learning algorithms provide an hypothesis-free, unbiased methodology, which can greatly enhance the search for predictors of therapeutic success in T2DM. The approach used in the present analysis provides an example of how a machine learning algorithm can be applied to a clinical dataset and used to develop predictions that can facilitate clinical decision making.
Plain Language Summary
What did this study look at?
-
This study looked at whether a computer program could predict which people with type 2 diabetes would respond best to a particular treatment.
-
The study treatment was a single-pill combination of two medicines, empagliflozin [em-PAH-gli-FLOW-zin] and linagliptin [LYNN-nah-GLIP-tin].
-
It is used to lower blood sugar (blood glucose) in people with type 2 diabetes.
-
-
The researchers used machine learning to analyze data from people who received this treatment. Machine learning uses computer models to find patterns in information.
-
The results helped to predict which people might respond best to the treatment.
Who took part in this study?
-
The researchers looked at results collected from two earlier studies of the treatment.
-
1363 people took part.
-
Approximately half of them were male.
-
Their average age was 55 years.
-
Approximately half of them had not received any previous diabetes treatment, and approximately half (50.3%) had received metformin treatment for diabetes.
-
What did the study show?
-
The researchers found that two blood tests commonly used in clinical practice helped them predict who would have the best response to treatment.
-
These tests were HbA1c levels (a measure of long-term blood glucose control), and their fasting plasma glucose (blood glucose levels when they had not eaten for 10–16 h).
-
-
This study suggests that machine learning could be a useful tool to help doctors decide which treatments will work best for individuals with type 2 diabetes.
Similar content being viewed by others
1 Introduction
Type 2 diabetes mellitus (T2DM) is a major cause of morbidity and mortality worldwide and its prevalence has been rising steadily over recent decades [1]. Although the number and availability of glucose-lowering agents have increased during recent years, the selection of appropriate treatment for individual patients with T2DM can be difficult given that the relative benefits/risks of different drugs for individual patients are not well understood [2], along with the limited knowledge about the association between patient characteristics and attainment of glycemic control. As common clinical practice indicates, to reach glycemic control the use of combination therapy with agents with complementary modes of action is frequently necessary [3]. The latest treatment strategy from the American Diabetes Association (ADA) recommends initial combination therapy when glycated hemoglobin (HbA1c) levels are ≥ 9% as this may provide more rapid attainment of HbA1c targets than with sequential therapy [4]. Similarly, the consensus statement by the American Association of Clinical Endocrinologists and American College of Endocrinology (AACE/ACE) states that dual combination therapy is usually required in patients with T2DM, and should be initiated when HbA1c is ≥ 7.5% [3]. However, despite these recommendations, glycemic control remains suboptimal in a significant proportion of patients [5,6,7], and availability of early predictors of glycemic response is lacking.
1.1 The Role of Machine Learning Techniques in Healthcare
Machine learning has recently been described as an important technology that can meaningfully process data that are beyond the capacity of the human brain to comprehend, in particular in relation to the huge clinical databases that are now available [8]. Although this remains a developing area in clinical medicine, a range of machine learning techniques are being increasingly used in healthcare, in particular to analyze the large and rapidly growing body of research and clinical data, and extract information that can lead to new hypotheses aimed at improved understanding and further investigation of medical conditions, including T2DM [9]. This approach can also be used to identify drug–target interactions in the search for potential candidates as a first step in the process of drug discovery [10]. To date, the main disease areas that have used machine learning techniques include oncology (e.g. for the prediction of breast cancer risk), cardiology (e.g. for predicting the occurrence of myocardial infarction), and neurology (e.g. in the evaluation of diagnostic imaging to predict outcomes after stroke) [11, 12]. In the field of diabetes, machine learning techniques have a range of applications, such as the use of computational algorithms in the evaluation of genomic data for the selection of biomarkers for T2DM [13], the identification of risk factors for predicting T2DM [14] or detecting individuals with impaired glucose tolerance or T2DM [15], for the prediction of T2DM following gestational diabetes [16], and in the classification of diabetic retinopathy [17]. An important limitation of machine learning models is that a model can only predict patient outcomes that are included in the dataset on which the model is based, and hence is dependent on the quality of data used to create the model [8]. Furthermore, the ease of interpretation of the model depends on the number of features evaluated—if the number is small, then the simple prediction tasks are easy to understand. In contrast, complex tasks are inherently more difficult to interpret because the model has been developed to identify complex statistical patterns, which might be more difficult to explain in terms of the subtle patterns that have led to a particular prediction [8]. It is important that clinicians who use machine learning systems understand how to interpret them so that they can identify clinical situations in which a model might be helpful. This underlines the need to perform real-world clinical evaluation of analytical models.
1.2 Application of Machine Learning to Clinical Datasets
The use of analytical methods to determine how treatments can benefit certain patients, and which patients will benefit from specific treatments, may help to improve treatment success in T2DM. Machine learning algorithms can be used as clinical prediction models to extract new information from the ever-increasing amounts of data generated by clinical trials. The machine learning algorithm provides an hypothesis-free, unbiased methodology that can facilitate the search for predictors of therapeutic success in T2DM. This approach can be used to find patterns in clinical datasets and offers the potential to define predictive factors to help identify which patients could benefit most from a given treatment [18]. This is particularly important in the field of diabetes research where predictors of response to antihyperglycemic therapies, both in terms of HbA1c reduction and maintenance of glycemic control, remain elusive. One type of machine learning involves the construction of computer systems that learn from experience to identify patterns in data and predict outcomes [19].
1.3 Description and Use of Random Forests
A commonly used type of machine learning methodology involves the use of random forests [20]. In this approach, decision trees derived from clinical data can be used to develop a group of trees (a random forest). A random forest is a group of tree-structured classifiers. The random forest approach incorporates two effective machine learning techniques—bagging and random feature selection. Bagging involves training each tree on a bootstrap sample of training data, and predictions are based on a majority vote of trees. This approach involves random selection of a subset of features to split at each node as a tree is grown. During training, each tree is grown using a particular bootstrap sample, with some of the data (approximately one-third) being left out during sampling. These omitted data are the out-of-bag (OOB) sample. Since the OOB data have not been used in tree construction, these data can be used to estimate the prediction performance [20]. Although each tree is unlikely to produce accurate predictions on its own, generating results based on the final vote across hundreds of trees can optimize the accuracy of predictions; the larger the number of trees, the greater the accuracy of the predictions [11, 20]. In the field of diabetes research, the random forest approach has the potential to explore relationships between possible disease predictors, and has been shown to help to screen potential biomarkers for T2DM [21]. This approach also has the potential to predict treatment success in T2DM by analyzing patient characteristics and treatment response.
1.4 Aims and Objectives
In the present study, clinical trial data comparing the single-pill combination of the sodium-glucose co-transporter-2/dipeptidyl peptidase-4 (SGLT2/DPP-4) inhibitor empagliflozin/linagliptin with empagliflozin or linagliptin monotherapies [22, 23] were used to determine if random forest or classification tree models could identify new predictors of treatment success, defined as HbA1c reduction. Specifically, the aims of this analysis were to identify patient characteristics associated with achieving an HbA1c target of ≤ 7% at week 12 and maintaining the target through week 52. By applying a new machine learning methodology to an existing clinical dataset, the practical application of this approach will be evaluated, and the potential utility of this new approach to clinical decision making can be assessed.
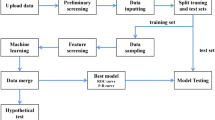
2 Methods
2.1 Design and Patients
Data were pooled from two phase III studies of empagliflozin/linagliptin single-pill combination therapy versus empagliflozin or linagliptin monotherapies in T2DM. These studies were chosen as a convenient sample that could be used to test the proposed analytical methods. The two studies had a similar design but enrolled patients who were treatment-naïve (study 1, n = 677) [22] or receiving background metformin (study 2, n = 686) [23]. Both trials were registered as NCT01422876 and have since been published, including a detailed description of the trial methods. In brief, both trials were randomized, double-blind, parallel-group studies that compared once-daily administration of a single-pill combination of empagliflozin plus linagliptin (empagliflozin 25 mg/linagliptin 5 mg, or empagliflozin 10 mg/linagliptin 5 mg) with empagliflozin monotherapy (25 mg or 10 mg daily) or linagliptin (5 mg) for 52 weeks. The inclusion criteria were patients aged ≥ 18 years, HbA1c level > 7% and ≤ 10.5% at screening, and a fasting plasma glucose (FPG) level of ≤ 240 mg/dL. Patients were randomized to one of five groups that were used for the descriptive analyses; however, for the machine learning data analysis, the two empagliflozin/linagliptin single-pill combination groups were pooled, as were the two empagliflozin monotherapy groups. In both studies, the primary endpoint was defined as the change in HbA1c levels between baseline and week 24.
2.2 Descriptive Analysis
Descriptive analysis was used to assess univariate associations between target categories and each baseline variable (e.g. HbA1c). Target attainment was defined by three groups: patients who achieved an HbA1c target of ≤ 7% at weeks 12 and 52; patients who reached the HbA1c target at week 12 but were above the target at week 52; and patients with HbA1c above the target at week 12 (irrespective of the week 52 value). Patients who discontinued before a specific time point were considered not at target for that time point. The differences in the distribution of at-target categories between groups were tested using a Chi-square test.
2.3 Machine Learning Analysis
Within the category of machine learning, the random forests algorithm is a well-established and now commonly used method. This method requires a dependent or outcome variable of interest and a list of independent variables as potential predictors of the outcome variable. The current study has a binary outcome variable (whether a patient had sustained response or not) and a relatively large set of patient characteristics as potential predictors, therefore a random forest approach was considered to be appropriate. The random forest algorithm was implemented using the randomForest R package (The R Foundation for Statistical Computing, Vienna, Austria).
After the descriptive analysis results, a machine learning analysis was planned and conducted (classification tree and random forest methods) to estimate and predict target categories based on patient characteristics at baseline without a priori selection. This analysis was based on the status at 12, 24, and 52 weeks. For stronger contrast, the analysis was limited to patients with sustained control (at target at all time points) or not in control (not at target at any time point). We excluded from the analysis patients with delayed control (not at target at week 12 but at target at either week 24 or 52) or non-sustained control (at target at week 12 but not at target at week 24 or 52).
Baseline variables included in the model were age, sex, race, ethnicity, geographic region, background therapy (treatment-naïve or receiving background metformin), height, weight, body mass index (BMI), waist circumference, smoking status, alcohol consumption, time since diagnosis, estimated creatinine clearance rate (CrCl), estimated glomerular filtration rate (eGFR), hypertension diagnosis, systolic blood pressure (SBP), diastolic blood pressure (DBP), HbA1c, and FPG.
2.4 Incorporation of the Random Forest Model
The random forest model is a well-established method in the statistical literature and has been increasingly applied in the field of biomedical research. In random forests, in general, the more trees, the better the prediction results. However, this improvement declines as the number of trees continues to grow, and, beyond a certain point, the amount of improvement becomes negligible. It has been shown in the random forest methodological literature that 500 is a sufficiently prudent number of trees to be built beyond which little improvement in prediction results is expected. Thus, in the present study, a conventional approach was used [24], with 500 individual trees constructed in each analysis; at each tree split, a random subset of 4 of 20 baseline variables were selected and considered as split candidates. The importance of the baseline variables was based on two parameters: (1) mean decrease in prediction accuracy without the variable in the model; and (2) mean decrease in the Gini index [25], a measure of impurity of the dataset (i.e. risk of misclassification of data), by including the variable. For both parameters, the greater the score, the greater the importance of the variable. An advantage of this approach is that the random forest method is robust in the presence of collinearity among potential predictors, unlike regression analysis.
For the present study, the descriptive analysis was performed using SAS 9.4 software (SAS Institute Inc., Cary, NC, USA); the machine learning analysis was performed in R version 3.3.2 (The R Project for Statistical Computing).
2.5 Use of Classification Tree Analysis as Comparator with Random Forest
Compared with the random forest method, the classification tree analysis is a simpler tree-based method that involves the construction of a single tree. In this study, it was used as a reference to compare with the random forest analysis. The two methods were compared using the training and validation set approach in which the full-analysis population was randomly divided into two subsets with a 60% versus 40% ratio (standard choice). The first subset (training set) was then used to build the models, and the second subset (validation set) was used to test the performance of the models.
3 Results
Overall, baseline patient characteristics were balanced between treatment groups, and the details have been published elsewhere for the individual studies [22, 23]. In summary, for the two studies overall (data given as mean ranges across study arms), the majority of participants were male (study 1, 48–58%; study 2, 46–61%), approximately 55 years of age (study 1, 53–56 years; study 2, 55–57 years), White (study 1, 70–78%; study 2, 71–76%), and diagnosed with T2DM at least 1–5 years previously (study 1, 36–45%; study 2, 34–37%). At baseline, mean HbA1c was approximately 8.0% in both studies (study 1, 7.99–8.05%; study 2, 7.90–8.02%) and mean FPG was approximately 156 mg/dL (study 1, 152.8–160.3 mg/dL; study 2, 154.6–161.6 mg/dL).
All treatment groups showed significant reductions from baseline in HbA1c over 24 weeks [22, 23]. Among patients who were treatment-naïve or receiving background metformin therapy, more of those treated with the empagliflozin/linagliptin single-pill combination achieved and maintained HbA1c targets compared with either agent alone (Table 1). The proportion of at-target categories in the single-pill combination groups was significantly greater than in the monotherapy groups (Chi-square test, p < 0.0001). In the descriptive analysis, lower mean baseline HbA1c and FPG were both associated with achieving and maintaining the HbA1c target (Table 1). Table 2 shows the number of patients who achieved sustained control in the trial, and the number predicted by the random forest model to achieve sustained glycemic control.
Figure 1 shows the performance of the random forest model across the three treatment groups. Overall, the graphs show the likelihood of error (y-axis) against the number of trees (x-axis). The rate of error decreases as the number of trees increases. The OOB estimates of the prediction error rate were 22.0%, 18.4%, and 22.5% for the empagliflozin/linagliptin, empagliflozin, and linagliptin analyses, respectively. Of the variables included in the model, baseline HbA1c and FPG were the two most important predictors (Fig. 2).
Prediction error rates for a empagliflozin/linagliptin single-pill combination, b empagliflozin, and c linagliptin. The graphs show the out-of-bag estimate of the prediction error rate (a measure of incorrect predictions); the prediction error rate among patients who achieved sustained glycemic control (analogous to the false-negative rate); and the prediction error rate among patients who did not achieve glycemic control (analogous to the false-positive rate)
Importance of baseline variables in the random forest analysis for a empagliflozin/linagliptin single-pill combination, b empagliflozin, and c linagliptin. BMI body mass index, CrCl estimated creatinine clearance rate, DBP diastolic blood pressure, eGFR estimated glomerular filtration rate, FPG fasting plasma glucose, HbA1c glycated hemoglobin, HTN hypertension, SBP systolic blood pressure, waist waist circumference
3.1 Machine Learning Analysis
The machine learning analysis also identified HbA1c and FPG as the strongest predictors of attaining glycemic control. As can be seen in Table 2, the error rate was related to the number of patients in each group (or pooled group). For example, in the empagliflozin/linagliptin group, 225 patients achieved sustained control and the model correctly predicted 194/225 patients (86.2%), but incorrectly predicted 31/225 patients (13.8%). Fewer patients were not in control throughout the study (n = 144), and the model incorrectly predicted that 50/144 patients (34.7%) would achieve sustained control.
3.2 Comparison with Classification Tree Analysis
In the validation set, small improvements in prediction accuracy for the random forest model versus the classification tree model were observed: 81% versus 79% for the empagliflozin/linagliptin single-pill combination, 82% versus 80% for empagliflozin, and 78% versus 77% for linagliptin.
4 Discussion
Using both traditional and novel data analysis methodologies, this study has identified baseline glycemic status as the strongest predictor of target glycemic control attainment. In the group of patients who received empagliflozin or linagliptin monotherapies or the empagliflozin/linagliptin single-pill combination, low baseline HbA1c and FPG (within the ranges evaluated in the two studies) predicted attainment of glycemic control (i.e. target HbA1c) during the treatment period. In contrast, covariates including body weight, waist circumference, SBP, DBP, or the other variables studied did not contribute to the outcome in the final model for any of the three therapies studied. These findings are consistent with current experience of glucose-lowering therapies [26,27,28], where high baseline HbA1c is associated with an increased HbA1c response to therapy but lower baseline HbA1c is associated with better achievement of target HbA1c since the initial HbA1c level is already close to the targeted value. HbA1c at the start of therapy has been shown to be predictive of HbA1c reductions achieved with insulin therapies [26], glucagon-like peptide-1 (GLP-1) agonists, DPP-4 inhibitors [27], metformin, and sulfonylureas [28].
While acknowledging several limitations of this study (as noted below) that might have compromised our ability to discover novel predictors of glycemic control attainment in this population, we report here similar results obtained with either an hypothesis-based or hypothesis-free data analysis methodology. This is consistent with the notion that the smaller the difference from target glycemic control, the more likely the success in reaching and maintaining glycemic control in response to a treatment. However, while this is a typically expected outcome, based on conventional hypothesis-driven analyses, the present study provides new insights into the use of a new machine learning algorithm to predict treatment responses. Furthermore, the magnitude of the influence of HbA1c and FPG, compared with the other evaluated variables, could provide important insights into the relative importance of known predictors of treatment response. A previous study also used a machine learning approach to identify predictors of treatment response to another SGLT2 inhibitor, dapagliflozin, and while expected predictors of treatment response were identified, the study demonstrated the potential utility of an hypothesis-independent approach in the evaluation of clinical data [18]. Since data from clinical trials are usually limited in the range of variables that can be evaluated due to the study design, future research could benefit from the use of real-world data sources, such as evaluation of datasets obtained from electronic health records. It is therefore possible that the use of such an hypothesis-free, unbiased methodology could be a useful way to enable the identification of baseline predictors of glycemic control and, in turn, to inform the choice of individualized, effective therapies for patients with diabetes. The machine learning algorithm used in our study is an example of how to approach this task. In particular, the random forest approach, as a machine learning method, offers the advantages of mimicking the human decision-making process and of providing personalized predictions for each assessed patient, for example, with respect to diagnosis, prognosis and treatment responses. These features make it an attractive tool to support clinicians in their practice and decision making [29].
The limitations of this study include the relatively small size of the population sample, which could restrict the generalizability of the findings. Furthermore, patients with delayed or non-sustained control were excluded from the analysis as the study was focused on the two extremes of response patterns (sustained control vs. not in control) with the aim of obtaining the greatest phenotypic contrast between these two patient groups to provide the best chance of detecting meaningful predictors of treatment response. The evaluation of patients with delayed or non-sustained control merits attention in future research. Another limitation of the study is that the set of variables measured at baseline in our study is limited and influenced by the design of clinical trials in T2DM. It is also possible that some predictive factors were not measured and were consequently not included in our analysis. Therefore, the prediction accuracy of the model could be improved by the inclusion of additional variables, such as T2DM biomarkers. The evaluation of larger and more comprehensive datasets, both in terms of the number of subjects and types of variables studied, is warranted. Real-world data sources, such as electronic health record data, are a promising alternative. The findings of this study suggest, however, that the use of hypothesis-free data analysis approaches is very promising and could have an important role in the search for predictors of therapeutic success in T2DM as this remains one of the most relevant criteria to guide therapeutic choices for patients with T2DM.
5 Conclusions
Identifying predictors of target glycemic control attainment can inform treatment choices and enhance success in treating diabetes. In this study, using both traditional and novel data analysis methodologies, we have identified baseline glycemic status as the strongest predictor of target glycemic control attainment. Machine learning algorithms provide an hypothesis-free, unbiased methodology, which can greatly enhance the search for predictors of therapeutic success in T2DM. We suggest that this approach may be applied to other drugs for which clinical datasets or real-world data are available. The approach used in the present analysis provides an example of how a machine learning algorithm can be applied to a clinical dataset and be used to develop predictions that can facilitate clinical decision making. The more widespread use of machine learning in healthcare has the potential to allow clinicians to take advantage of medically relevant data and assist them in the provision of optimal and individualized patient care.
References
World Health Organization. Global report on diabetes. 2016. http://apps.who.int/iris/bitstream/10665/204871/1/9789241565257_eng.pdf. Accessed 14 Mar 2017.
Ma Q, Lu AY. Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacol Rev. 2011;63(2):437–59.
Garber AJ, Abrahamson MJ, Barzilay JI, et al. Consensus Statement by the American Association of Clinical Endocrinologists and American College of Endocrinology on the comprehensive type 2 diabetes management algorithm—2019 executive summary. Endocr Pract. 2019;25(1):69–100.
American Diabetes Association. 9. Pharmacologic approaches to glycemic treatment: standards of medical care in diabetes—2019. Diabetes Care. 2019;42(Suppl 1):S90–S102.
Fox KM, Gerber RA, Bolinder B, Chen J, Kumar S. Prevalence of inadequate glycemic control among patients with type 2 diabetes in the United Kingdom general practice research database: a series of retrospective analyses of data from 1998 through 2002. Clin Ther. 2006;28(3):388–95.
Khan H, Lasker SS, Chowdhury TA. Exploring reasons for very poor glycaemic control in patients with type 2 diabetes. Prim Care Diabetes. 2011;5(4):251–5.
Stark Casagrande S, Fradkin JE, Saydah SH, Rust KF, Cowie CC. The prevalence of meeting A1C, blood pressure, and LDL goals among people with diabetes, 1988–2010. Diabetes Care. 2013;36(8):2271–9.
Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019;380(14):1347–58.
Kavakiotis I, Tsave O, Salifoglou A, Maglaveras N, Vlahavas I, Chouvarda I. Machine learning and data mining methods in diabetes research. Comput Struct Biotechnol J. 2017;15:104–16.
Chen R, Liu X, Jin S, Lin J, Liu J. Machine learning for drug-target interaction prediction. Molecules. 2018;23(9):E2208.
Deo RC. Machine learning in medicine. Circulation. 2015;132(20):1920–30.
Jiang F, Jiang Y, Zhi H, et al. Artificial intelligence in healthcare: past, present and future. Stroke Vasc Neurol. 2017;2(4):230–43.
Cai L, Wu H, Li D, Zhou K, Zou F. Type 2 diabetes biomarkers of human gut microbiota selected via iterative sure independent screening method. PLoS One. 2015;10(10):e0140827.
Lee BJ, Kim JY. Identification of type 2 diabetes risk factors using phenotypes consisting of anthropometry and triglycerides based on machine learning. IEEE J Biomed Health Inform. 2016;20(1):39–46.
Hische M, Luis-Dominguez O, Pfeiffer AF, Schwarz PE, Selbig J, Spranger J. Decision trees as a simple-to-use and reliable tool to identify individuals with impaired glucose metabolism or type 2 diabetes mellitus. Eur J Endocrinol. 2010;163(4):565–71.
Wang KJ, Adrian AM, Chen KH, Wang KM. An improved electromagnetism-like mechanism algorithm and its application to the prediction of diabetes mellitus. J Biomed Inform. 2015;54:220–9.
Casanova R, Saldana S, Chew EY, Danis RP, Greven CM, Ambrosius WT. Application of random forests methods to diabetic retinopathy classification analyses. PLoS One. 2014;9(6):e98587.
Bujac S, Del Parigi A, Sugg J, et al. Patient characteristics are not associated with clinically important differential response to dapagliflozin: a staged analysis of phase 3 data. Diabetes Ther. 2014;5(2):471–82.
Anderson JP, Parikh JR, Shenfield DK, et al. Reverse engineering and evaluation of prediction models for progression to type 2 diabetes: an application of machine learning using electronic health records. J Diabetes Sci Technol. 2016;10(1):6–18.
Breiman L. Random forests. Machine Learning. 2001;45:5–32.
Huang JH, He RH, Yi LZ, Xie HL, Cao DS, Liang YZ. Exploring the relationship between 5’AMP-activated protein kinase and markers related to type 2 diabetes mellitus. Talanta. 2013;110:1–7.
Lewin A, DeFronzo RA, Patel S, et al. Initial combination of empagliflozin and linagliptin in subjects with type 2 diabetes. Diabetes Care. 2015;38(3):394–402.
DeFronzo RA, Lewin A, Patel S, et al. Combination of empagliflozin and linagliptin as second-line therapy in subjects with type 2 diabetes inadequately controlled on metformin. Diabetes Care. 2015;38(3):384–93.
Hastie TTR, Friedman J. The elements of statistical learning: data mining, inference, and prediction. 2nd ed. Berlin: Springer; 2009.
Strobl CBA, Augustin T. Unbiased split selection for classification trees based on the Gini Index. Comput Stat Data Anal. 2007;52:483–501.
Balkau B, Calvi-Gries F, Freemantle N, Vincent M, Pilorget V, Home PD. Predictors of HbA1c over 4 years in people with type 2 diabetes starting insulin therapies: the CREDIT study. Diabetes Res Clin Pract. 2015;108(3):432–40.
Bihan H, Ng WL, Magliano DJ, Shaw JE. Predictors of efficacy of GLP-1 agonists and DPP-4 inhibitors: a systematic review. Diabetes Res Clin Pract. 2016;121:27–34.
Martono DP, Lub R, Lambers Heerspink HJ, Hak E, Wilffert B, Denig P. Predictors of response in initial users of metformin and sulphonylurea derivatives: a systematic review. Diabet Med. 2015;32(7):853–64.
Kavakiotis I, Tsave O, Salifoglou A, Maglaveras N, Vlahavas I, Chouvarda I. Machine learning and data mining methods in diabetes research. Comput Struct Biotechnol J. 2017;15:104–16.
Acknowledgements
The authors meet the criteria for authorship as recommended by the International Committee of Medical Journal Editors (ICMJE) and were fully responsible for all content and editorial decisions, were involved at all stages of manuscript development, and approved the final version that reflects the authors’ interpretations and conclusions.
Data Availability
The datasets generated and/or analyzed during the current study are available in the Clinical Study Data Request repository (https://clinicalstudydatarequest.com/).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
The studies were sponsored by the Boehringer Ingelheim and Eli Lilly and Company Diabetes Alliance; this analysis was supported by Boehringer Ingelheim Pharmaceuticals, Inc. (BIPI). The authors received no direct compensation related to the development of the manuscript. Writing support was provided by Jennifer Garrett, MBBS, of Envision Scientific Solutions, which was contracted and compensated by BIPI for this service. Open access was funded by BIPI.
Conflict of Interest
Richard Pratley has received research funding from Lexicon Pharmaceuticals, Lilly, Merck, Novo Nordisk, Sanofi Aventis US, LLC, and Takeda; speaker fees from AstraZeneca, Boehringer Ingelheim, Novo Nordisk, and Takeda; and consultancy fees from AstraZeneca, Boehringer Ingelheim, Janssen Scientific Affairs, LLC, Ligand Pharmaceuticals, Inc., Lilly, Merck, Novo Nordisk, Sanofi Aventis US, LLC, and Takeda. All honoraria and fees are directed to a non-profit organization; he received no direct compensation. Dacheng Liu and Wenbo Tang are employees of Boehringer Ingelheim. Angelo Del Parigi and Christopher Lee were employees of Boehringer Ingelheim at the time these studies were conducted.
Ethical Approval
In the studies analyzed, all procedures performed involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed Consent
Informed consent was obtained from all individual participants included in the studies.
Prior Publication
This work was previously presented as a poster at the American Association of Clinical Endocrinologists (AACE) 26th Annual Scientific and Clinical Congress, Austin, TX, USA, 3–7 May 2017.
Additional information
Enhanced Digital Features
To view enhanced digital features for this article go to https://doi.org/10.6084/m9.figshare.7999478.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Del Parigi, A., Tang, W., Liu, D. et al. Machine Learning to Identify Predictors of Glycemic Control in Type 2 Diabetes: An Analysis of Target HbA1c Reduction Using Empagliflozin/Linagliptin Data. Pharm Med 33, 209–217 (2019). https://doi.org/10.1007/s40290-019-00281-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40290-019-00281-4