Abstract
Introduction
Iatrogeny due to drug–drug interactions is insufficiently documented, due to the high number of possible combinations.
Objective
This study aimed to design a simple but general method to predict the variation of adverse events (AE) frequency due to a pharmacokinetic or pharmacodynamic interaction.
Methods
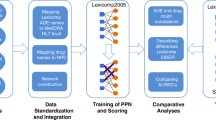
Three prediction models were designed using a logistic probability density function. Each prediction model was based on three components: the AE odds ratio of each drug in the combination, and the area under the curve ratio (Rauc) of the pharmacokinetic interaction, if any. Pharmacodynamic interaction was assumed to be additive on logit scale. Rauc was predicted using a well-validated mechanistic static model, freely available online. No combination study is required. The method was evaluated against a wide range of AEs (28 High Level Terms) and 211 drug combinations (involving 43 victim drugs and 55 perpetrators), by comparing the observed and predicted frequencies. The observed odds ratios were estimated with a disproportionality analysis from the FDA Adverse Event Reporting System, using an approach that minimizes biases.
Results
With the best model, the rate of prediction considered as correct (within 50–200% of the observed value) was 72%, and the bias was negligible (-5%). The AE odds ratio due to pharmacokinetic and pharmacodynamic interactions was equally well predicted.
Conclusions
A simple workflow to implement the method in practice is proposed. This method may help to foresee and to anticipate the harmful consequences associated with drug–drug interactions, at virtually no experimental cost, when the odds ratio of an AE is known for each drug alone and the AUC ratio is known or predicted by a suitable model.





Similar content being viewed by others
References
Laatikainen O, Miettunen J, Sneck S, Lehtiniemi H, Tenhunen O, Turpeinen M. The prevalence of medication-related adverse events in inpatients-a systematic review and meta-analysis. Eur J Clin Pharmacol. 2017;73(12):1539–49. https://www.ncbi.nlm.nih.gov/pubmed/28871436.
Gonzaga de Andrade Santos TN, Mendonca da Cruz Macieira G, Cardoso Sodre Alves BM, Onozato T, Cunha Cardoso G, Ferreira Nascimento MT, et al. Prevalence of clinically manifested drug interactions in hospitalized patients: a systematic review and meta-analysis. PLoS One. 2020;15(7):e0235353. https://www.ncbi.nlm.nih.gov/pubmed/32609783.
Dechanont S, Maphanta S, Butthum B, Kongkaew C. Hospital admissions/visits associated with drug–drug interactions: a systematic review and meta-analysis. Pharmacoepidemiol Drug Saf. 2014;23(5):489–97. https://www.ncbi.nlm.nih.gov/pubmed/24616171.
Momo K, Kobayashi H, Sugiura Y, Yasu T, Koinuma M, Kuroda SI. Prevalence of drug–drug interaction in atrial fibrillation patients based on a large claims data. PLoS One. 2019;14(12):e0225297. https://www.ncbi.nlm.nih.gov/pubmed/31815956.
Szarfman A, Machado SG, O'Neill RT. Use of screening algorithms and computer systems to efficiently signal higher-than-expected combinations of drugs and events in the US FDA's spontaneous reports database. Drug Saf. 2002;25(6):381–92. https://www.ncbi.nlm.nih.gov/pubmed/12071774.
Almenoff JS, DuMouchel W, Kindman LA, Yang X, Fram D. Disproportionality analysis using empirical Bayes data mining: a tool for the evaluation of drug interactions in the post-marketing setting. Pharmacoepidemiol Drug Saf. 2003;12(6):517–21. https://www.ncbi.nlm.nih.gov/pubmed/14513665.
Tatonetti NP, Ye PP, Daneshjou R, Altman RB. Data-driven prediction of drug effects and interactions. Sci Transl Med. 2012;4(125):125ra31. https://www.ncbi.nlm.nih.gov/pubmed/22422992.
Administration USFaD. U.S. Food and Drug Administration Guidance. In vitro drug interaction studies—cytochrome P450 enzyme- and transporter-mediated drug interactions (2020).
Administration USFaD. U.S. Food and Drug Administration Guidance. 2020. Clinical drug interaction studies—cytochrome P450 enzyme- and transporter-mediated drug interactions (2020).
Minto C, Vuyk J. Response surface modelling of drug interactions. Adv Exp Med Biol. 2003;523:35–43. https://www.ncbi.nlm.nih.gov/pubmed/15088838.
Wicha SG, Chen C, Clewe O, Simonsson USH. A general pharmacodynamic interaction model identifies perpetrators and victims in drug interactions. Nat Commun. 2017;8(1):2129. https://www.ncbi.nlm.nih.gov/pubmed/29242552.
Ohno Y, Hisaka A, Suzuki H. General framework for the quantitative prediction of CYP3A4-mediated oral drug interactions based on the AUC increase by coadministration of standard drugs. Clin Pharmacokinet. 2007;46(8):681–96. https://www.ncbi.nlm.nih.gov/pubmed/17655375.
Castellan AC, Tod M, Gueyffier F, Audars M, Cambriels F, Kassai B, et al. Quantitative prediction of the impact of drug interactions and genetic polymorphisms on cytochrome P450 2C9 substrate exposure. Clin Pharmacokinet. 2013;52(3):199–209. https://www.ncbi.nlm.nih.gov/pubmed/23344982.
Gabriel L, Tod M, Goutelle S. Quantitative prediction of drug interactions caused by CYP1A2 inhibitors and inducers. Clin Pharmacokinet. 2016;55(8):977–90. https://www.ncbi.nlm.nih.gov/pubmed/26936044.
Goutelle S, Bourguignon L, Bleyzac N, Berry J, Clavel-Grabit F, Tod M. In vivo quantitative prediction of the effect of gene polymorphisms and drug interactions on drug exposure for CYP2C19 substrates. AAPS J. 2013;15(2):415–26. https://www.ncbi.nlm.nih.gov/pubmed/23319287.
Tod M, Goutelle S, Clavel-Grabit F, Nicolas G, Charpiat B. Quantitative prediction of cytochrome P450 (CYP) 2D6-mediated drug interactions. Clin Pharmacokinet. 2011;50(8):519–30. https://www.ncbi.nlm.nih.gov/pubmed/21740075.
Tod M, Goutelle S, Gagnieu MC, Genophar IIWG. Genotype-based quantitative prediction of drug exposure for drugs metabolized by CYP2D6. Clin Pharmacol Ther. 2011;90(4):582–7. https://www.ncbi.nlm.nih.gov/pubmed/21866098.
Tod M, Nkoud-Mongo C, Gueyffier F. Impact of genetic polymorphism on drug–drug interactions mediated by cytochromes: a general approach. AAPS J. 2013;15(4):1242–52. https://www.ncbi.nlm.nih.gov/pubmed/24027036.
Open FDA's API. https://open.fda.gov/. Accessed 15 Apr 2021.
Letinier L, Ferreira A, Marceron A, Babin M, Micallef J, Miremont-Salame G, et al. Spontaneous reports of serious adverse drug reactions resulting from drug–drug interactions: an analysis from the french pharmacovigilance database. Front Pharmacol. 2020;11:624562. https://www.ncbi.nlm.nih.gov/pubmed/33841134.
Open FDA's fields definition. https://open.fda.gov/apis/drug/event/searchable-fields/. Accessed 15 Apr 2021.
FDA's Orange Book. https://www.fda.gov/media/76860/download. Accessed 15 Apr 2021.
OHDSI vocabulary. https://athena.ohdsi.org/vocabulary/list. Accessed 15 Apr 2021.
DrugBank. https://www.drugbank.com/. Accessed 15 Apr 2021.
Banda JM, Evans L, Vanguri RS, Tatonetti NP, Ryan PB, Shah NH. A curated and standardized adverse drug event resource to accelerate drug safety research. Sci Data. 2016;3:160026. https://www.ncbi.nlm.nih.gov/pubmed/27193236
Harada T, Kosaka S, Elliesen J, Yasuda M, Ito M, Momoeda M. Ethinylestradiol 20 mug/drospirenone 3 mg in a flexible extended regimen for the management of endometriosis-associated pelvic pain: a randomized controlled trial. Fertil Steril. 2017;108(5):798–805. https://www.ncbi.nlm.nih.gov/pubmed/28911925.
FDA Adverse Event Reporting System (FAERS) Public Dashboard. https://www.fda.gov/drugs/questions-and-answers-fdas-adverse-event-reporting-system-faers/fda-adverse-event-reporting-system-faers-public-dashboard. Accessed 18 Apr 2022.
Hazell L, Shakir SA. Under-reporting of adverse drug reactions: a systematic review. Drug Saf. 2006;29(5):385–96. https://www.ncbi.nlm.nih.gov/pubmed/16689555.
Teng C, Frei CR. Delirium associations with antibiotics: a pharmacovigilance study of the FDA adverse event reporting system (FAERS). Drugs Real World Outcomes. 2022;9(1):23–9. https://www.ncbi.nlm.nih.gov/pubmed/34275113.
Khouri C, Petit C, Tod M, Lepelley M, Revol B, Roustit M, et al. Adverse drug reaction risks obtained from meta-analyses and pharmacovigilance disproportionality analyses are correlated in most cases. J Clin Epidemiol. 2021;134:14–21. https://www.ncbi.nlm.nih.gov/pubmed/33508405.
King G, Nielsen R. Why propensity scores should not be used for matching. Polit Anal. 2019;27(4):435–54.
Fermier N, Bourguignon L, Goutelle S, Bleyzac N, Tod M. Identification of cytochrome P450-mediated drug–drug interactions at risk in cases of gene polymorphisms by using a quantitative prediction model. Clin Pharmacokinet. 2018;57(12):1581–91. https://www.ncbi.nlm.nih.gov/pubmed/29572664.
Simon F, Gautier-Veyret E, Truffot A, Chenel M, Payen L, Stanke-Labesque F, et al. Modeling approach to predict the impact of inflammation on the pharmacokinetics of CYP2C19 and CYP3A4 substrates. Pharm Res. 2021;38(3):415–28. https://www.ncbi.nlm.nih.gov/pubmed/33686560.
Savic RM, Mentre F, Lavielle M. Implementation and evaluation of the SAEM algorithm for longitudinal ordered categorical data with an illustration in pharmacokinetics-pharmacodynamics. AAPS J. 2011;13(1):44–53. https://www.ncbi.nlm.nih.gov/pubmed/21063925.
Acknowledgements
The authors thank Ms. Gaelle SIMEON for language editing of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No funding was received for this work.
Conflict of interest
The authors declared no competing interests for this work.
Data availability
The database used for this study is available from the corresponding author on reasonable request.
Author contributions
MT and MA wrote the manuscript; MT designed the research; MT and TR performed the research. All authors read and approved the final version.
Ethical approval
Not applicable since no patients were enrolled and the data are publicly available.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Code availability
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tod, M., Rodier, T. & Auffret, M. Quantitative Prediction of Adverse Event Probability Due to Pharmacokinetic Interactions. Drug Saf 45, 755–764 (2022). https://doi.org/10.1007/s40264-022-01190-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40264-022-01190-3