Abstract
Background and Objectives
Ganciclovir (GCV) and valganciclovir (VGCV) show large interindividual pharmacokinetic variability, particularly in children. The objectives of this study were (1) to develop machine learning (ML) algorithms trained on simulated pharmacokinetics profiles obtained by Monte Carlo simulations to estimate the best ganciclovir or valganciclovir starting dose in children and (2) to compare its performances on real-world profiles to previously published equation derived from literature population pharmacokinetic (POPPK) models achieving about 20% of profiles within the target.
Materials and Methods
The pharmacokinetic parameters of four literature POPPK models in addition to the World Health Organization (WHO) growth curve for children were used in the mrgsolve R package to simulate 10,800 pharmacokinetic profiles. ML algorithms were developed and benchmarked to predict the probability to reach the steady-state, area-under-the-curve target (AUC0–24 within 40–60 mg × h/L) based on demographic characteristics only. The best ML algorithm was then used to calculate the starting dose maximizing the target attainment. Performances were evaluated for ML and literature formula in a test set and in an external set of 32 and 31 actual patients (GCV and VGCV, respectively).
Results
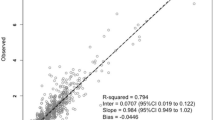
A combination of Xgboost, neural network, and random forest algorithms yielded the best performances and highest target attainment in the test set (36.8% for GCV and 35.3% for the VGCV). In actual patients, the best GCV ML starting dose yielded the highest target attainment rate (25.8%) and performed equally for VGCV with the Franck model formula (35.3% for both).
Conclusion
The ML algorithms exhibit good performances in comparison with previously validated models and should be evaluated prospectively.






Similar content being viewed by others
References
Zuhair M, Smit GSA, Wallis G, Jabbar F, Smith C, Devleesschauwer B, et al. Estimation of the worldwide seroprevalence of cytomegalovirus: a systematic review and meta-analysis. Rev Med Virol. 2019;29: e2034.
Hayes M, Newman AM, Boge CLK, Galetaki DM, Elgarten CW, Freedman JL, et al. Incidence of CMV infection and disease and adverse events associated with antiviral therapy in a retrospective cohort of allogeneic hematopoietic cell transplant recipients at an academic children’s hospital. J Pediatr Infect Dis Soc. 2021;10:910–8.
Downes KJ, Sharova A, Boge CLK, Vader D, Mitrou M, Hayes M, et al. CMV infection and management among pediatric solid organ transplant recipients. Pediatr Transpl. 2022;26: e14220.
Manuel O, Kralidis G, Mueller NJ, Hirsch HH, Garzoni C, van Delden C, et al. Impact of antiviral preventive strategies on the incidence and outcomes of cytomegalovirus disease in solid organ transplant recipients. Am J Transpl. 2013;13:2402–10.
Kotton CN, Kumar D, Caliendo AM, Huprikar S, Chou S, Danziger-Isakov L, et al. The third international consensus guidelines on the management of cytomegalovirus in solid-organ transplantation. Transplantation. 2018;102:900–31.
Rastogi S, Ricci A, Jin Z, Bhatia M, George D, Garvin JH, et al. Clinical and economic impact of cytomegalovirus infection among children undergoing allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transpl. 2019;25:1253–9.
Bateman CM, Kesson A, Powys M, Wong M, Blyth E. Cytomegalovirus infections in children with primary and secondary immune deficiencies. Viruses. 2021;13:2001.
Franck B, Autmizguine J, Marquet P, Ovetchkine P, Woillard J-B. Pharmacokinetics, pharmacodynamics, and therapeutic drug monitoring of valganciclovir and ganciclovir in transplantation. Clin Pharmacol Ther. 2022;112:233–76.
Padullés A, Colom H, Bestard O, Melilli E, Sabé N, Rigo R, et al. Contribution of population pharmacokinetics to dose optimization of ganciclovir-valganciclovir in solid-organ transplant patients. Antimicrob Agents Chemother. 2016;60:1992–2002.
Peled O, Berkovitch M, Rom E, Bilavsky E, Bernfeld Y, Dorfman L, et al. Valganciclovir dosing for cytomegalovirus prophylaxis in pediatric solid-organ transplant recipients: a prospective pharmacokinetic study. Pediatr Infect Dis J. 2017;36:745–50.
Jorga K, Reigner B, Chavanne C, Alvaro G, Frey N. Pediatric dosing of ganciclovir and valganciclovir: how model-based simulations can prevent underexposure and potential treatment failure. CPT Pharmacometr Syst Pharmacol. 2019;8:167–76.
Ponthier L, Ensuque P, Destere A, Marquet P, Labriffe M, Jacqz-Aigrain E, et al. Optimization of vancomycin initial dose in term and preterm neonates by machine learning. Pharm Res. 2022;39:2497–506.
Yao B-F, Wu Y-E, Tang B-H, Hao G-X, Jacqz-Aigrain E, van den Anker J, et al. Predictive performance of pharmacokinetic model-based virtual trials of vancomycin in neonates: mathematics matches clinical observation. Clin Pharmacokinet. 2022;61:1027–38.
Woillard J-B, Labriffe M, Prémaud A, Marquet P. Estimation of drug exposure by machine learning based on simulations from published pharmacokinetic models: the example of tacrolimus. Pharmacol Res. 2021;167: 105578.
Franck B, Woillard J-B, Théorêt Y, Bittencourt H, Demers E, Briand A, et al. Population pharmacokinetics of ganciclovir and valganciclovir in paediatric solid organ and stem cell transplant recipients. Br J Clin Pharmacol. 2021;87:3105–14.
Schwartz GJ, Brion LP, Spitzer A. The use of plasma creatinine concentration for estimating glomerular filtration rate in infants, children, and adolescents. Pediatr Clin North Am. 1987;34:571–90.
Elmokadem A, Riggs MM, Baron KT. Quantitative systems pharmacology and physiologically-based pharmacokinetic modeling with mrgsolve: a hands-on tutorial. CPT Pharmacometrics Syst Pharmacol. 2019;8:883–93.
Nguyen T, Oualha M, Briand C, Bendavid M, Béranger A, Benaboud S, et al. Population pharmacokinetics of intravenous ganciclovir and oral valganciclovir in a pediatric population to optimize dosing regimens. Antimicrob Agents Chemother. 2021;65:e02254-e2320.
Facchin A, Elie V, Benyoub N, Magreault S, Maisin A, Storme T, et al. Population pharmacokinetics of ganciclovir after valganciclovir in renal transplant children. Antimicrob Agents Chemother. 2019;63:e01192-19, AAC.01192-19.
Razonable RR, Humar A. Cytomegalovirus in solid organ transplant recipients–guidelines of the American society of transplantation infectious diseases community of practice. Clin Transplant. 2019;33: e13512.
Labriffe M, Woillard J-B, Debord J, Marquet P. Machine learning algorithms to estimate everolimus exposure trained on simulated and patient pharmacokinetic profiles. CPT Pharmacometr Syst Pharmacol. 2022;11:1018.
Kuhn M, Wickham H. tidymodels: Easily Install and Load the ‘Tidymodels’ Packages version 1.1.0 from CRAN. [Internet]. https://rdrr.io/cran/tidymodels/
Chen T, Guestrin C. XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining [Internet]. San Francisco California USA: ACM; 2016 [cited 2021 Jun 30]. p. 785–94. https://doi.org/10.1145/2939672.2939785.
Cheng B, Titterington DM. Neural networks: a review from a statistical perspective. Stat Sci [Internet]. 1994. https://doi.org/10.1214/ss/1177010638.full.
Breiman L. No title found. Mach Learn [Internet]. 2001;45:5–32. https://doi.org/10.1023/A:1010933404324.
Couch S, Kuhn M. Tidy Model Stacking Version 1.0.2. https://stacks.tidymodels.org/
Pescovitz MD, Ettenger RB, Strife CF, Sherbotie JR, Thomas SE, McDiarmid S, et al. Pharmacokinetics of oral valganciclovir solution and intravenous ganciclovir in pediatric renal and liver transplant recipients. Transpl Infect Dis. 2010;12:195–203.
Åsberg A, Bjerre A, Neely M. New algorithm for valganciclovir dosing in pediatric solid organ transplant recipients. Pediatr Transplant. 2014;18:103–11.
Bououda M, Uster DW, Sidorov E, Labriffe M, Marquet P, Wicha SG, et al. A machine learning approach to predict interdose vancomycin exposure. Pharm Res. 2022;39:721–31.
Uster DW, Stocker SL, Carland JE, Brett J, Marriott DJE, Day RO, et al. A model averaging/selection approach improves the predictive performance of model-informed precision dosing: vancomycin as a case study. Clin Pharmacol Ther. 2021;109:175–83.
Smits A, Kulo A, N de Hoon J, Allegaert K. Pharmacokinetics of drugs in neonates: pattern recognition beyond compound specific observations. CPD [Internet]. 2012 [cited 2022 Jan 4];18:3119–46. http://www.eurekaselect.com/openurl/content.php?genre=article&issn=1381-6128&volume=18&issue=21&spage=3119
Biccler JL, Eloranta S, de Nully BP, Frederiksen H, Jerkeman M, Jørgensen J, et al. Optimizing outcome prediction in diffuse large b-cell lymphoma by use of machine learning and nationwide lymphoma registries: a Nordic lymphoma group study. JCO Clin Cancer Inform. 2018;2:1–13.
Shakhovska N, Yakovyna V, Chopyak V. A new hybrid ensemble machine-learning model for severity risk assessment and post-COVID prediction system. Math Biosci Eng. 2022;19:6102–23.
Acknowledgements
The authors thanks Karen Poole for manuscript editing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
The authors have no competing interests to declare in relation to this work.
Funding information
No funding was received for this study.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Data availability
The datasets generated during and/or analyzed during the current study are not publicly available due to privacy and ethical restrictions but are available from the corresponding author on reasonable request.
Data transparency
All the values of covariates and the code used to simulate them are available at https://github.com/ponthL/ganciclovir_first_dose.git.
Author contributions
LP, JBW, PM, ML contributed to the study conception and design. Material preparation, data collection and analysis were performed by BF, AA, JA, PO. AD developed the shiny app. The first draft of the manuscript was written by LP and JBW and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ponthier, L., Autmizguine, J., Franck, B. et al. Optimization of Ganciclovir and Valganciclovir Starting Dose in Children by Machine Learning. Clin Pharmacokinet 63, 539–550 (2024). https://doi.org/10.1007/s40262-024-01362-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40262-024-01362-7