Abstract
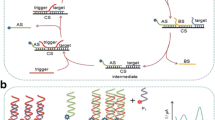
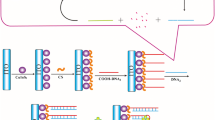
Target detection circuits have been previously designed, which are propelled by conventional PCR, isothermal amplification and strand-displacement reaction. These detection circuits obtain the target signal via the replication of the target strand, the aggregation of the signal particles or the branch migration. Here we constructed a triode-like enzyme-free catalyst strand-displacement circuit for target DNA detection. The target strand triggered the reaction and released the fluorescence signal strand circularly through branch migration. However, the main challenge of strand-displacement reaction is the signal leakage. Therefore, we designed a double strand structure “junction fuel”, which was used to increase the binding energy across the displacement process. Ultimately, the leakage of the system obtained stable inhabitation due to the junction fuel strand. The limit of detection of the system was as low as 0.11 nmol/L and the gain of the system was as high as 28-fold(the concentration of target was 50 nmol/L). Furthermore, the process of the system was visualized vividly in the reaction curve through the kinetic simulation implemented, which suggests that the combination of the kinetic simulation and the experiment exhibits a promising prospect towards the use of strand-displacement circuit in analytical, diagnostic application and synthetic biology.
Similar content being viewed by others
References
Yurke B., Andrew J. T., Allen P. M. J., Nature, 2000, 406(2000), 605
Simmel F. C., Yurke B., Singh H. R., Chem. Rev., 2019, 119(10), 6326
Song J., Li Z., Wang P., Meyer T., Mao C., Ke Y., Science, 2017, 357(2017), 3377
Wang D., Song J., Wang P., Pan V., Zhang Y., Cui D., Ke Y., Nat. Protoc., 2018, 13(10), 2312
Fan S., Chen J., Ji B., Gao C., Jiang K., Liu Y., Song J., Chinese Science Bulletin, 2018, 64(10), 1027
Fan S., Wang D., Kenaan A., Cheng J., Cui D., Song J., Small, 2019, 15(26), 1805554
Chen W., He B., Li C., Zhang X., Wu W., Yin X., Fan B., Fan X., Wang J., J. Med. Microbiol., 2007, 56(2007), 603
Zhao Y., Chen F., Li Q., Wang L., Fan C., Chem. Rev., 2015, 115(22), 12491
Wang X., Yan N., Song T., Wang B., Wei B., Lin L., Chen X., Tian H., Liang H., Advanced Biosystems, 2017, 1(6), 1700060
Ho N. R. Y., Lim G. S., Sundah N. R., Lim D., Loh T. P., Shao H., Nat. Commun., 2018, 9(1), 3238
Sun X., Wei B., Guo Y., Xiao S., Li X., Yao D., Yin X., Liu S., Liang H., J. Am. Chem. Soc., 2018, 140(31), 9979
Fern J., Scalise D., Cangialosi A., Howie D., Potters L., Schulman R., ACS Synth Biol., 2017, 6(2), 190
Seelig G., Soloveichik. D., Zhang. D. Y., Winfree E., Science, 2006, 314(2006), 1585
Karunanayake M. A., Yu Q., Leon-Duque M. A., Zhao B., Wu R., You M., J. Am. Chem. Soc., 2018, 140(28), 8739
Shi C., Liu Q., Ma C., Zhong W., Anal. Chem., 2014, 86(1), 336
Zhao Y., Zhou L., Tang Z., Nat. Commun., 2013, 4(2013), 1493
Song T., Eshra A., Shah S., Bui H., Fu D., Yang M., Mokhtar R., Reif J., Nat. Nanotechnol., 2019, 14(11), 1075
Rothemund P. W., Nature, 2006, 440(7082), 297
Ji B., Song J., Wang D., Kenaan A., Zhu Q., Wang J., Sonderskov S. M., Dong M., Langmuir, 2019, 35(11), 4140
Thubagere A. J., Li W., Johnson R. F., Chen Z., Doroudi S., Lee Y. L., Izatt G., Wittman S., Srinivas N., Woods D., Winfree E., Qian L., Science, 2017, 357(6356), No. eaan6558
Deng H., Liu Q., Wang X., Huang R., Liu H., Lin Q., Zhou X., Xing D., Biosens. Bioelectron., 2017, 87(2017), 931
Zhu W., Su X., Gao X., Dai Z., Zou X., Biosens. Bioelectron., 2014, 53(2014), 414
Wang J., Jiang X., Han H., Biosens. Bioelectron., 2016, 82(2016), 26
Ma Z. Y., Ruan Y. F., Xu F., Zhao W. W., Xu J. J., Chen H. Y., Anal. Chem., 2016, 88(7), 3864
Song T., Liang H., J. Am. Chem. Soc., 2012, 134(26), 10803
Song T., Xiao S., Yao D., Huang F., Hu M., Liang H., Adv. Mater., 2014, 26(35), 6181
Tavallaie R., McCarroll J., Le Grand M., Ariotti N., Schuhmann W., Bakker E., Tilley R. D., Hibbert D. B., Kavallaris M., Gooding J. J., Nat. Nanotechnol., 2018, 13(11), 1066
Wei X., Zhou W., Sanjay S. T., Zhang J., Jin Q., Xu F., Dominguez D. C., Li X., Anal. Chem., 2018, 90(16), 9888
Canoura J., Wang Z. W., Yu H. X., Alkhamis O., Fu F. F., Xiao Y., J. Am. Chem. Soc., 2018, 140(31), 9961
Ramlal S., Mondal B., Lavu P. S., N B., Kingston J., Int. J. Food Microbiol, 2018, 265(2018), 74
Song Y., Shi Y., Huang M., Wang W., Wang Y., Cheng J., Lei Z., Zhu Z., Yang C., Angew. Chem. Int. Ed. Engl., 2019, 58(8), 2236
Xiong X., Shi X., Liu Y., Lu L., You J., Analytical Methods, 2018, 10(3), 365
Yang J., Jiang S. X., Liu X. R., Pan L. Q., Zhang C., ACS Applied Materials & Interfaces, 2016, 8(49), 34054
Wang S. S., Ellington A. D., Chem. Rev., 2019, 119(10), 6370
Frank-Kamenetskii M., Nature, 1987, 328(6125), 17
Qiao Y., Qian Y., Liu M., Liu N., Tang X., Chem. Res. Chinese Universities, 2019, 35(5), 837
Li B., Ellington A. D., Chen X., Nucleic. Acids Res., 2011, 39(16), No. e100
Shi K., Dou B., Yang C., Chai Y., Yuan R., Xiang Y., Anal. Chem., 2015, 87(16), 8578
Watson J. D., Crick J. D., Nature, 1953, 171(1953), 737
Qian L., Winfree E., Bruck J., Nature, 2011, 475(7356), 368
Qian L., Winfree E., Science, 2011, 332(6034), 1196
Gao Z., Xia H., Zauberman J., Tomaiuolo M., Ping J., Zhang Q., Ducos P., Ye H., Wang S., Yang X., Lubna F., Luo Z., Ren L., Johnson A. T. C., Nano Lett., 2018, 18(6), 3509
Zhang C., Wang Z., Liu Y., Yang J., Zhang X., Li Y., Pan L., Ke Y., Yan H., J. Am. Chem. Soc., 2019, 141(43), 17189
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by the National Key Research and Development Program of China(No.2017YFC1200904), the National Natural Science Foundation of China(Nos.11761141006, 21605102) and the Natural Science Foundation of Shanghai, China (Nos.19520714100, 19ZR1475800).
Supporting Information
Rights and permissions
About this article
Cite this article
Luo, T., Wang, X., Fan, S. et al. A Triode-like Enzyme-free Catalytic Circuit with Junction Fuel. Chem. Res. Chin. Univ. 36, 261–267 (2020). https://doi.org/10.1007/s40242-019-0025-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40242-019-0025-2