Abstract
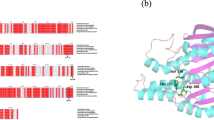
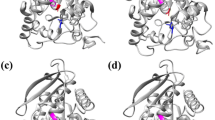
Better understanding of the relationship between the substrate preference and structural module of esterases is helpful to novel enzyme development. For this purpose, two chimeric esterases AAM7 and PAR, constructed via domain swapping between two ancient thermophilic esterases, were investigated on their molecular simulation(including homology modeling, substrates docking and substrate binding affinity validation) and enzymatic assay(specific activities and activation energies calculating). Our results indicate that the factors contributing to the substrate preference of many enzymes especially the broad-specificity enzymes like esterases are multiple and complicated, the substrate binding domains or binding pockets are important but not the only factor for substrate preference.
Similar content being viewed by others
References
Bornscheuer U. T., Curr. Opin. Biotechnol., 2002, 13, 543
Panda T., Gowrishankar B. S., Appl. Micorbiol. Biotechnol., 2005, 67, 160
Osterlund T., Eur. J. Biochem., 2001, 268, 1899
Bornscheuer U. T., Bessler C., Srinivas R., Krishna S. H., Trends Biotechnol., 2002, 20, 433
Nardini M., Dijkstra B. W., Curr. Opin. Struct. Biol., 1999, 9, 732
Holmquist M., Curr. Protein Pept. Sci., 2000, 1, 209
Manco G., Giosuè E., D’Auria S., Herman P., Carrea G., Rossi M., Arch. Biochem. Biophys., 2000, 373, 182
Gao R., Feng Y., Ishikawa K., Ishida H., Ando S., Kosugi Y., Cao S., J. Mol. Catal. B, Enzym., 2003, 24, 1
de Simone G., Menchise V., Manco G., Mandrich L., Sorrentino N., Lang D., Rossi M., Pedone C., J. Mol. Biol., 2001, 314, 507
Bartlam M., Wang G., Yang H., Gao R., Zhao X., Xie G., Cao S., Feng Y., Rao Z., Structure, 2004, 12, 1481
Tao J., Zhao B., Tian X., Zheng L., Cao S., Chem. Res. Chinese Universities, 2010, 26(5), 816
Thompson J. D., Higgins D. G., Gibson T. J., Nucleic Acids Res., 1994, 22, 4673
DeLano W. L., The PyMOL Molecular Graphics System (Version 1.5.0.4 Schrodinger, LLC), 2002, http://www.pymol.org
Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A., Curr. Protoc. Protein Sci., Chapter 2, Unit 2.9, John Wiley & Sons, Inc., New York, 2007
Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M., J. App. Cryst., 1993, 26, 283
Wang Y., Wang Y., Han W., Feng Y., Chem. Res. Chinese Universities, 2012, 28(4), 707
Huey R., Morris G. M., Olson A. J., Goodsell D. S., J. Comput. Chem., 2007, 28, 1145
Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J., J. Comput. Chem., 2009, 30, 2785
Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., Olson A. J., J. Comput. Chem., 1998, 19, 1639
Yang G., Bai A., Gao L., Feng Y., Biochim. Biophys. Acta, 2009, 1794, 94
Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J., Nucleic Acids Res., 2006, 34, W116
Zhou X., Wang H., Zhang Y., Gao L., Feng Y., Acta Biochim. Biophys. Sin., 2012, 44, 965
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supported by the National Basic Research Program of China(Nos.2012CB721000, 2011CBA00800) and the National Natural Science Foundation of China(No.30970632).
Rights and permissions
About this article
Cite this article
Zhou, Xl., Han, Ww., Zheng, Bs. et al. Insight into substrate preference of two chimeric esterases by combining experiment and molecular simulation. Chem. Res. Chin. Univ. 29, 533–537 (2013). https://doi.org/10.1007/s40242-013-2353-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40242-013-2353-y