Abstract
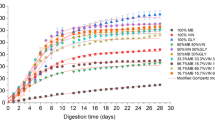
A consortium of bacteria capable of decomposing oily hydrocarbons was isolated from tarballs on the beaches of Terengganu, Malaysia, and classified as Pseudomonas stutzeri, Cellulosimicrobium cellulans, Acinetobacter baumannii and Pseudomonas balearica. The Taguchi design was used to optimize the biodegradation of diesel using these bacteria as a consortium. The highest biodegradation of diesel-oil in the experimental tests was 93.6%, and the individual n-alkanes decomposed 87.6—97.6% over 30 days. Optimal settings were inoculum size of 2.5 mL (1.248 OD600nm); 12% (v/v) the initial diesel-oil in a minimal salt medium of pH 7.0, 30.0 gL−1 NaCl and 2.0 gL−1 NH4NO3 concentration, incubated at 42 °C temperature and 150 rpm agitation speed. Parameters significantly improved diesel-oil removal by consortium as shown by the model determination coefficient (R2 = 90.89%; P < 0.001) with a synergistic effect of agitation speed significantly contributing 81.03%. Taguchi design determined the optimal settings for the parameters under study, which significantly improved diesel-oil removal by consortium. This can be used to design a novel bioremediation strategy that can achieve optimal decontamination of oil pollution in a shorter time.
Highlights
-
Hydrocarbon-degraders in Tarball were isolated and identified by their 16S rRNA gene sequence as Pseudomonas stutzeri, Cellulosimicrobium cellulans, Acinetobacter baumannii and Pseudomonas balearica.
-
Taguchi method was applied to optimize effects of parameters such as initial diesel concentration, salinity (NaCl concentration), nitrate (NH4NO3) concentration, pH, temperature, agitation speed and inoculum size on diesel-oil removal in 30 days.
-
Maximum diesel-oil biodegradation by experimental runs was 93.6% with individual n-alkanes degraded between 87.6% – 97.6% in 30 days
-
Optimal settings were 2.5 mL (1.248 OD600nm) inoculum size; 12% (v/v) initial diesel-oil in MSM media with 7.0 pH, 30.0 gL−1 NaCl and 2.0 gL−1 NH4NO3 concentration, incubated at 42 °C temperature and 150 rpm agitation speed
-
Parameters significantly improved diesel-oil removal by this consortium as indicated by model determination coefficient (R2 = 90.89%; P < 0.001) with synergistic effect of agitation speed significantly contributing 81.03%
-
Taguchi design established optimal settings of investigated parameters that produced significant improvement on diesel-oil removal by consortium







Similar content being viewed by others
Data availability
We further confirm that datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- µL:
-
Microliter
- 16S rRNA:
-
16S ribosomal RNA
- A. baumannii :
-
Acinetobacter baumannii
- Adj. SS:
-
Adjusted sum of squares
- Adj. MS:
-
Adjusted mean squares
- ANOVA:
-
Analysis of Variance
- BLAST:
-
Basic Local Alignment System Tool
- °C:
-
Degree Celsius
- C. cellulans :
-
Cellulosimicrobium cellulans
- CI:
-
Confidence interval
- conc.:
-
Concentration
- DCM:
-
Dichloromethane
- df:
-
Degree of freedom
- DNA:
-
Deoxyribonucleic Acid
- dNTP:
-
Deoxy nucleoside triphosphates
- g:
-
Gram
- GC-MS:
-
Gas Chromatography Mass Spectrometry
- gL− 1 :
-
Gram per litre
- h:
-
Hours
- HPLC:
-
High performance liquid chromatography
- L:
-
Liter
- LB:
-
Luria Bertani
- MEGA:
-
Molecular Evolutionary Genetics Analysis
- mg:
-
Milligram
- mL:
-
Milliliter
- MSM:
-
Minimal salt media
- NaCl:
-
Sodium chloride
- NB:
-
Nominal is best
- NCBI:
-
National Center for Biotechnology Information
- ng/mg:
-
Nanogram per milligram
- NH4NO3 :
-
Ammonium nitrate
- OD600nm :
-
Optical density at 600 nm
- OFAT:
-
One-factor-a-time
- P. balearica :
-
Pseudomonas balearica
- PCR:
-
Polymerase chain reaction
- ppm:
-
Parts per million
- P. stutzeri :
-
Pseudomonas stutzeri
- rpm:
-
Revolutions per minute
- SE Coefficients:
-
Standard error coefficients
- secs:
-
Seconds
- SIS:
-
Surrogate internal standard
- SN:
-
Signal noise
- v/v:
-
Volume per volume
References
Abatenh E, Gizaw B, Tsegaya Z, Wassie M. Application of microorganisms in bioremediation-review. J Environ Microbiol. 2017;1(1):2–9.
Abbasian F, Lockington R, Mallavarapu M, Naidu R. A comprehensive review of aliphatic hydrocarbon biodegradation by bacteria. Appl Biochem Biotechnol. 2015;176:670–99.
Adzigbli L, Yuewen D. Assessing the impact of oil spills on marine organisms. J Oceanogr. 2018;3(6):179.
Agarry SE. Statistical optimization and kinetic studies of enhanced bioremediation of crude oil - contaminated marine water using combined adsorption-biostimulation strategy. J Appl Sci Environ Manag. 2017;21(1):59.
Aghamiri SF, Kabiri K, Emtiazi G. A novel approach for optimization of crude oil bioremediation in soil by the taguchi method. J Pet Environ Biotechnol. 2011;2(2):1–7.
Al-Sayegh A, Al-Wahaibi Y, Joshi S, Al-Bahry S, Elshafie A, Al-Bemani A. Bioremediation of heavy crude oil contamination. Open Biotechnol J. 2016;10(1):301–11.
Amer R, Abdel-Fattah YR. Hydrocarbonoclastic marine bacteria in Mediterranean Sea, El-Max, Egypt: isolation, identification and site characterization. Jokull J. 2014;64(4):42–58.
Armstrong JA, Schulz JR. Agarose Gel Electrophoresis. Current Protocols in Essential Laboratory Techniques 2015; 2015. 721-7222.
Avanzi IR, Gracioso LH, Baltazar MDPG, Karolski B, Perpetuo EA, Nascimento CAO. Aerobic biodegradation of gasoline compounds by bacteria isolated from a hydrocarbon-contaminated soil. Environ Eng Sci. 2015;32(12):990–7.
Ayed BH, Jemil N, Maalej H, Bayoudh A, Hmidet N, Nasri M. Enhancement of solubilization and biodegradation of diesel oil by biosurfactant from Bacillus amyloliquefaciens An6. Int Biodeterior Biodegradation. 2015;99:8–14.
Azubuike CC, Chikere CB, Okpokwasili GC. Bioremediation techniques–classification based on site of application: principles advantages limitations and prospects. World J Microbiol Biotechnol. 2016;32(11):1–18.
Bacosa HP, Thyng KM, Plunkett S, Erdner DL, Liu Z. The tarballs on Texas beaches following the 2014 Texas City “Y” Spill: Modeling, chemical, and microbiological studies. Mar Pollut Bull. 2016;109(1):236–44.
Baharudzaman EH, Tuah PM. Distribution and abundance of stranded tarballs in Marintaman Beach, Sipitang, Sabah, Malaysia. Int J Adv Res Sci Eng Technol. 2018;5(3):5396–400.
Baird RB, Eaton AD, Rice EW. American Water Works Association and Water Environment Federation In Standard Methods for the Examination of Water and Wastewater. 23rd ed. Washington: American Public Health Association; 2017. 2017.
Bellas J, Saco-Alvarez L, Nieto O, Bayona JM, Albaiges J, Beiras R. Evaluation of artificially-weathered standard fuel oil toxicity by marine invertebrate embryogenesis bioassays. Chemosphere. 2013;90(3):1103–8.
Bennasar-Figueras A, Salvà-Serra F, Jaén-Luchoro D, Seguí C, Aliaga F, Busquets A, Lalucat J. Complete genome sequence of Pseudomonas balearica DSM 6083T. Genome Announc. 2016;4(2):1–2.
Bhattacharya M, Biswas D, Guchhait S. Applied growth kinetic models for crude oil spill bioremediation in a batch scale bioreactor. J Environ Hazards. 2018;1(1):1–6.
Bio-Rad. PCR troubleshooting. Bio-Rad - Life Sci Res. 2016;2016:1–22.
Blackburn M, Mazzacano CAS, Fallon C, Black SH. Oil in our oceans: A review of the impacts of oil spills on marine invertebrates. Portland, Oregon. The Xerces Society for Invertebrate Conservation. 2014. 2014: 152.
Borah D, Yadav RNS. Bioremediation of petroleum based contaminants with biosurfactant produced by a newly isolated petroleum oil degrading bacterial strain. Egypt J Pet. 2017;26(1):181–8.
Borowik A, Wyszkowska J. Remediation of soil contaminated with diesel oil. J Elementol. 2018;23(2):767–88.
Brown LD, Cologgi DL, Gee KF, Ulrich AC Bioremediation of Oil Spills on Land. In Oil Spill Science and Technology. 2nd ed. 2017. 2017: 699–729.
Brzeszcz J, Kaszycki P. Aerobic bacteria degrading both n-alkanes and aromatic hydrocarbons: an undervalued strategy for metabolic diversity and flexibility. Biodegradation. 2018;29:359–407.
Cappuccino JG, Welsh C. Microbiology. A Laboratory Manual. Pearson Education Limited. 11th ed. 2017. 2017.
Catania V, Cappello S, Di Giorgi V, Santisi S, Di Maria R, Mazzola A, Quatrini P. Microbial communities of polluted sub-surface marine sediments. Mar Pollut Bull. 2018;131:396–406.
Challa S, Neelapu NRR. Phylogenetic Trees: Applications Construction and Assessment In K R Hakeem N A Shaik B Banaganapalli and R Elango (Eds). Essentials of Bioinformatics Volume III: In Silico Life Sciences: Agriculture. Cham: Springer International Publishing; 2019. 2019: 167–192.
Chen Q, Li J, Liu M, Sun H, Bao M. Study on the biodegradation of crude oil by free and immobilized bacterial consortium in marine environment. PLoS One. 2017;12(3):1–23.
Coskun O. Separation techniques: chromatography. North Clin Istanb. 2016;3(2):156–60.
Costa JC, Oliveira JV, Alves MM. Response surface design to study the influence of inoculum particle size and inoculum to substrate ratio on the methane production from Ulex sp. Renew Energy. 2016;96(B):1071–7.
Daghio M, Tatangelo V, Franzetti A, Gandolfi I, Papacchini M, Careghini A, Bestetti G. Hydrocarbon degrading microbial communities in bench scale aerobic biobarriers for gasoline contaminated groundwater treatment. Chemosphere. 2015;130:34–9.
Dashtbozorg M, Riyahi Bakhtiari A, Shushizadeh MR, Taghavi L. Quantitative evaluation of n-alkanes PAHs and petroleum biomarker accumulation in beach-stranded tarballs and coastal surface sediments in the Bushehr Province Persian Gulf (Iran). Mar Pollut Bull. 2019;146:801–15.
Dashti N, Ali N, Eliyas M, Khanafer M, Sorkhoh NA, Radwan SS. Most hydrocarbonoclastic bacteria in the total environment are diazotrophic which highlights their value in the bioremediation of hydrocarbon contaminants. Microbes Environ. 2015;30:70–5.
Deng MCC, Li J, Liang FRR, Yi M, Xu XMM, Yuan JPP, Wang JHH. Isolation and characterization of a novel hydrocarbon-degrading bacterium Achromobacter sp HZ01 from the crude oil-contaminated seawater at the Daya Bay southern China. Mar Pollut Bull. 2014;83:79–86.
Dong L, Lv LB, Lai R. Molecular Cloning A laboratory Manual. 4th ed. 2012. 33: 11–115.
Dou TY, Luan HW, Ge GB, Dong MM, Zou HF, He YQ, Yang L. Functional and structural properties of a novel cellulosome-like multienzyme complex: efficient glycoside hydrolysis of water-insoluble 7-xylosyl-10-deacetylpaclitaxel. Sci Rep. 2015;5:13768.
Drabik A, Bodzoń-Kułakowska A, Silberring J. Gel Electrophoresis. In Proteomic Profiling and Analytical Chemistry: The Crossroads. 2nd ed. 2016. 2016: 115–143.
Environment Canada HC. Final Screening Assessment Report for Pseudomonas stutzeri. Pseudomonas Stutzeri ATCC 17587. 2015. 2015: 1–40.
Eppendorf,. OD600 measurements using different photometers. White Paper. 2015;28:1–4.
Farag S, Soliman NA, Abdel-Fattah YR. Statistical optimization of crude oil bio-degradation by a local marine bacterium isolate Pseudomonas sp SP48. J Genet Eng Biotechnol. 2018;16(2):409–20.
Ferreira TF, Coelho MAZ, da Rocha-Leão MHM. Factors influencing crude oil biodegradation by Yarrowia lipolytica. Braz Arch Biol Technol. 2012;55(5):785–91.
Fingas MF. The basics of oil spill cleanup. 3rd ed. Boca Raton: CRC Press; 2013. 2013: 225.
Fu Y, Cheng L, Meng Y, Li S, Zhao X, Du Y, Yin H. Cellulosimicrobium cellulans strain E4–5 enzymatic hydrolysis of curdlan for production of (1 → 3)-linked β-d-glucan oligosaccharides. Carbohyd Polym. 2015;134:740–4.
Gao YC, Wang JN, Guo SH, Hu YL, Li TT, Mao R, Zeng DH. Effects of salinization and crude oil contamination on soil bacterial community structure in the Yellow River Delta region China. Appl Soil Ecol. 2015;86:165–73.
Gopinath LR, Divya D, Geitha TR, Bhuvaneswari R, Archaya S, Merlin CP. Hydrocarbon degradation and biogas production efficiency of bacteria isolated from petrol polluted soil. Res J Recent Sci. 2015;4(9):60–7.
Hamzah A, Manikan V, Abd Aziz NAF. Biodegradation of tapis crude oil using consortium of bacteria and fungi: Optimization of crude oil concentration and duration of incubation by response surface methodology. Sains Malaysiana. 2017;46(1):43–50.
Hassanshahian M, Abarian M, Cappello S. Biodegradation of aromatic compounds biodegradation and bioremediation of polluted systems. New Adv Technol. 2015;6:110–8.
Ibrahim M, Makky EA, Azmi NS, Ismail J. Optimization parameters for Mycobacteria confluentis biodegradation of PAHs. MATEC Web Conf. 2018;06035(150):1–5.
Ikner L, Schmitz B, Gerba C, Pepper I. Bacterial growth curve analysis and its environmental applications. JoVe. 2015;2015:1–11.
Imron MF, Titah HS. Optimization of diesel biodegradation by Vibrio alginolyticus using Box-Behnken design. Environ Eng Res. 2018;23(4):374–82.
Islam B. Petroleum sludge its treatment and disposal: a review. Int J Chem Sci. 2015;13(4):1584–602.
Jiang Y, Qi H, Zhang X. Novel method for separation and screening of lubricant-degrading microorganisms and bacterial biodegradation. Chin J Chem Eng. 2016;24(3):353–9.
Kaczorek E, Bielicka-Daszkiewicz K, Héberger K, Kemény S, Olszanowski A, Voelkel A. Best conditions for biodegradation of diesel oil by chemometric tools. Braz J Microbiol. 2014;45(1):117–26.
Kamaruzzaman JA, Zain AM. Coral Bay shore zones tarball distribution. Procedia Eng. 2016;148:437–43.
Kaur R, Kumari A, Kaur R. Microbial degradation of crude-petroleum-oil: factors and strategies affecting the bioremediation process. Pollut Res. 2018;37(4):1053–7.
Krasowska A, Sigler K. How microorganisms use hydrophobicity and what does this mean for human needs. Front Cell Infect Microbiol. 2014;4:112.
Kumar V, Kumar M, Prasad R. Microbial degradation of hydrocarbons in the environment: an overview. Microbial Action Hydrocarbons. 2019;2019:353–86.
Lee PY, Costumbrado J, Hsu CY, Kim YH. Agarose gel electrophoresis for the separation of DNA fragments. J Vis Exp. 2012;62(e3923):1–5.
Lee CR, Lee JH, Park M, Park KS, Bae IK, Kim YB, Lee SH. Biology of Acinetobacter baumannii: Pathogenesis antibiotic resistance mechanisms and prospective treatment options. Front Cell Infect Microbiol. 2017;7(55):1–35.
Liu Z, Liu J. Evaluating bacterial community structures in oil collected from the sea surface and sediment in the northern Gulf of Mexico after the Deepwater Horizon oil spill. Microbiol Open. 2013;2(3):492–504.
Liu L, Mishchenko MI. Modeling study of scattering and absorption properties of tar-ball aggregates. Appl Opt. 2019;58(31):8648–57.
Liu S, Wang B, Wang Y. Identification of Diesel Residues by GC / MS / MS. International Conference on New Energy and Renewable Resources. 2017. 2: 341–345.
Liu H, Xu J, Liang R, Liu J. Characterization of the medium- And long-chain n-alkanes degrading Pseudomonas aeruginosa strain SJTD-1 and its alkane hydroxylase genes. PLoS One. 2014;9(8):1–14.
Liu B, Ju M, Liu J, Wu W, Li X. Isolation identification and crude oil degradation characteristics of a high-temperature hydrocarbon-degrading strain. Mar Pollut Bull. 2016;106(1–2):301–7.
McGenity TJ, Timmis KN, Nogales FB. Hydrocarbon and Lipid Microbiology Protocols. Activities and Phenotypes. Springer Protocols Handbooks; 2017;2017: 7-67.
Messaoudene NA. Taguchi design of experiments. Analysis. 2010;2010:71–6.
Mishra PR, Gupta EN, Joshi D. Prediction of moulding sand properties using multiple regression methodology. J Adv Comput Commun Technol. 2016;4(1):1–4.
Mukherjee AK, Bhagowati P, Biswa BB, Chanda A, Kalita B. A comparative intracellular proteomic profiling of Pseudomonas aeruginosa strain ASP-53 grown on pyrene or glucose as sole source of carbon and identification of some key enzymes of pyrene biodegradation pathway. J Proteomics. 2017;167:25–35.
Muthukamalam S, Sivagangavathi S, Dhrishya D, Sudha Rani S. Characterization of dioxygenases and biosurfactants produced by crude oil degrading soil bacteria. Braz J Microbiol. 2017;48(4):637–47.
Mwaura AN. Screening and characterization of hydrocarbonoclastic bacteria isolated from oil-contaminated soils from auto garages. Int J Microbiol Biotechnol. 2018;3(1):1–11.
Nazifa TH, Ahmad MAB, Hadibarata T, Salmiati S, Aris A. Bioremediation of diesel oil spill by filamentous fungus Trichoderma reesei H002 in aquatic environment. Int J Integrated Eng. 2018;10(9):14–9.
Nkem BM, Halimoon N, Yusoff FM, Johari WLW, Zakaria MP, Medipally SR, Kannan N. Isolation identification and diesel-oil biodegradation capacities of indigenous hydrocarbon-degrading strains of Cellulosimicrobium cellulans and Acinetobacter baumannii from tarball at Terengganu beach Malaysia. Mar Pollut Bull. 2016;107(1):261–8.
Nkem BM, Halimoon N, Yusoff FM, Johari WLW. Isolation and optimization of diesel-oil biodegradation using Cellulosimicrobium cellulans from tarball. Pertanika J Sci Technol. 2019;27(3):1031–40.
Notowidjaja MSI, Ekawati Y, Noya S. Taguchi experimental design to optimize the sugar content of candied carrot. IOP Conf Ser Mater Sci Eng. 2019;528: 012068.
Omotoyinbo O. Effect of Varying NaCl Concentrations on the Growth Curve of Escherichia coli and Staphylococcus aureus. Cell Biol. 2016;4(5):31–4.
Palanisamy N, Ramya J, Kumar S, Vasanthi NS, Chandran P, Khan S. Diesel biodegradation capacities of indigenous bacterial species isolated from diesel contaminated soil. J Environ Health Sci Eng. 2014;12(1):1–8.
Pandey P, Pathak H, Dave S. Biodegradation of diesel oil by Pseudomonas balearica strain UKMS3P3 isolated from soil around mathura refinery. Indian J Environ Prot. 2018;38(6):467–76.
Paniagua-Michel J, Fathepure BZ. Microbial consortia and biodegradation of petroleum hydrocarbons in marine environments. Microbial Action Hydrocarbons. 2019;2019:1–20.
Parthipan P, Elumalai P, Sathishkumar K, Sabarinathan D, Murugan K, Benelli G, Rajasekar A. Biosurfactant and enzyme mediated crude oil degradation by Pseudomonas stutzeri NA3 and Acinetobacter baumannii MN3. 3 Biotech. 2017;7(5):1–1.
Payne JR, Phillips CR. Tarball Formation and Distribution. Petroleum Spills in the Marine Environment Boca Raton CRC Press. 1st ed. 2018. 2018: 83–97.
Pereira MR, Mercaldi GF, Maester TC, Balan A, De Macedo Lemos EG. Est16 a new esterase isolated from a metagenomic library of a microbial consortium specializing in diesel oil degradation. PLoS One. 2015;10(7): e0133723.
Pereira TM, Merçon J, Passos LS, Coppo GC, Lopes TOM, Cabral DS, Chippari-Gomes AR. Effects of the water-soluble fraction of diesel oil (WSD) on the fertilization and development of a sea urchin (Echinometra lucunter). Ecotoxicol Environ Saf. 2018;162:59–62.
Polmear R, Stark JS, Roberts D, McMinn A. The effects of oil pollution on Antarctic benthic diatom communities over 5 years. Mar Pollut Bull. 2015;90(1–2):33–40.
Pundir R, Chary GHVC, Dastidar MG. Application of Taguchi method for optimizing the process parameters for the removal of copper and nickel by growing Aspergillus sp. Water Resour Ind. 2018;20:83–92.
Qin W, Fan F, Zhu Y, Huang X, Ding A, Liu X, Dou J. Anaerobic biodegradation of benzo(a)pyrene by a novel Cellulosimicrobium cellulans CWS2 isolated from polycyclic aromatic hydrocarbon-contaminated soil. Braz J Microbiol. 2018;49(2):258–68.
Rajabnasab M, Khavari-Nejad RA, Shokravi S, Nejadsattari T. Investigating the physiological responses of three endaphic strains of Cyanobacteria to crude oil concentrations in limited salinity and irradiation conditions. Appl Ecol Environ Res. 2018;16(4):4559–73.
Ramasamy S, Arumugam A, Chandran P. Optimization of Enterobacter cloacae (KU923381) for diesel oil degradation using response surface methodology. J Microbiol. 2017;55(2):104–11.
Rao S, Samant P, Kadampatta A, Shenoy R. An Overview of Taguchi Method: Evolution Concept and Interdisciplinary Applications. Int J Sci Eng Res. 2013;4(10):621–6.
Reddy CM, Arey JS, Seewald JS, Sean PS, Lemkau KL, Nelson RK, Carmichael CA, McIntyre CP, Fenwick J, Ventura GT, Van Mooy BAS, Camilli R. Composition and fate of gas and oil released to the water column during the Deepwater Horizon oil spill. Proc Natl Acad Sci USA. 2012;109:20229–34.
Rengarajan T, Rajendran P, Nandakumar N, Lokeshkumar B, Rajendran P, Nishigaki I. Exposure to polycyclic aromatic hydrocarbons with special focus on cancer. Asian Pac J Trop Biomed. 2015;5(3):182–9.
Roslee AFA, Zakaria NN, Convey P, Zulkharnain A, Lee GLY, Gomez-Fuentes C, Ahmad SA. Statistical optimisation of growth conditions and diesel degradation by the Antarctic bacterium Rhodococcus sp strain AQ5-07. Extremophiles. 2019;24(2):277–91.
Roy A, Dutta A, Pal S, Gupta A, Sarkar J, Chatterjee A, Kazy SK. Biostimulation and bioaugmentation of native microbial community accelerated bioremediation of oil refinery sludge. Biores Technol. 2018;253:22–32.
Sadeghi HZM, Ebrahimipour G, Shahriari MM, Fakhari J, Abdoli T. Bioremediation of crude oil using bacterium from the coastal sediments of Kish Island Iran. Iran J Public Health. 2016;45(5):670–9.
Sawadogo A, Otoidobiga HC, Nitiema LW, Traoré AS, Dianou D. Optimization of hydrocarbons biodegradation by bacterial strains isolated from wastewaters in ouagadougou burkina faso: case study of SAE 40/50 Used Oils and Diesel. J Agric Chem Environ. 2016;5(1):1–11.
Shaieb FM, Elghazawani AH, Issa A. Studies on crude oil degrading Bacteria isolated from Libyan desert. Int J Curr Microbiol App Sci. 2015;4(2):920–7.
Shine H, Samant LR, Tulaskar V, Vartak D. Isolation of potent hydrocarbon degrading microorganisms and its application in bioremediation. Int J Curr Pharm Res. 2017;9(3):65.
Shirneshan G, Bakhtiari AR, Memariani M. Identification of sources of tarballs deposited along the Southwest Caspian Coast, Iran using fingerprinting techniques. Sci Total Environ. 2016;568:979–89.
Singh P, Parmar D, Pandya A. Parametric optimization of media for the crude oil degrading bacteria isolated from crude oil contaminated site. Int J Curr Microbiol App Sci. 2015;4(2):322–8.
Sivagamasundari T, Jayakumar N. Optimization of diesel oil degrading bacterial strains at various culture parameters. Int J Res Dev Pharm Life Sci. 2017;6(6):2840–4.
Stagars MH, Emil Ruff S, Amann R, Knittel K. High diversity of anaerobic alkane-degrading microbial communities in marine seep sediments based on (1-methylalkyl) succinate synthase genes. Front Microbiol. 2016;6(1511):1–17.
Stauffer E. Gas Chromatography-Mass Spectrometry. Encyclopedia of Forensic Sciences Elsevier Inc. 2nd ed. 2013;2013: 596–602.
Suneel V, Vethamony P, Naik BG, Krishna MS, Jadhav L. Identifying the source of tarballs deposited along the beaches of Goa in 2013 and comparing with historical data collected along the West Coast of India. Sci Total Environ. 2015;528:313–21.
Tan YH, Chiang EW, Chan HYL, Ling S, Hii, Kit K, Woo. Behavioral properties of Acinetobacter species in degrading oil. Malays J Biochem Mol Biol. 2018;21(3):72–6.
Tanzadeh J, Ghasemi MF. the Use of Microorganisms in Bioremediation of Oil Spills in Sea Waters and Shoreline. Chem Biol Interface. 2016;5(6):282–9.
Tripathi NK, Shrivastava A. Scale up of biopharmaceuticals production. Nanoscale Fabrication Optimization Scale-up and Biological Aspects of Pharmaceutical Nanotechnology. 2017. 2017: 133–172.
Uba BO, Chukwura EI, Okoye EL, Ubani O, Chude CO, Akabueze UC. In vitro degradation and reduction of aromatic hydrocarbons by marine bacteria isolated from contaminated marine environments of Niger delta. Adv Res. 2019;18(3):1–17.
Umar ZD, Nor Azwady AA, Zulkifli SZ, Muskhazli M. Effective phenanthrene and pyrene biodegradation using Enterobacter sp MM087 (KT933254) isolated from used engine oil contaminated soil. Egypt J Pet. 2018;27(3):349–59.
Urakawa H, Rajan S, Feeney ME, Sobecky PA, Mortazavi B. Ecological response of nitrification to oil spills and its impact on the nitrogen cycle. Environ Microbiol. 2019;21(1):18–33.
Valdor PF, Gómez AG, Puente A. Environmental risk analysis of oil handling facilities in port areas application to Tarragona harbor (NE Spain). Mar Pollut Bull. 2015;90:78–87.
Varjani SJ, Gnansounou E, Pandey A. Comprehensive review on toxicity of persistent organic pollutants from petroleum refinery waste and their degradation by microorganisms. Chemosphere. 2017;188:280–91.
Vulin C, Leimer N, Huemer M, Ackermann M, Zinkernagel AS. Prolonged bacterial lag time results in small colony variants that represent a sub-population of persisters. Nat Commun. 2018;9:1–7.
Wood JL, Osman A, Wade SA. An efficient cost-effective method for determining the growth rate of sulfate-reducing bacteria using spectrophotometry. Methods X. 2019;6:2248–57.
Woodman ME, Savage CR, Arnold WK, Stevenson B. Direct PCR of intact bacteria (colony PCR). Curr Protocols Microbiol. 2016;2016:A3D1-A3D7.
Wu Y, Zeng J, Zhu Q, Zhang Z, Lin X. PH is the primary determinant of the bacterial community structure in agricultural soils impacted by polycyclic aromatic hydrocarbon pollution. Sci Rep. 2017;7(40093):1–7.
Yetti E, A’La A, Mercuriani IS, Yopi Y. Potency of microbial consortium F14 from three selected bacterial strains (Labrenzia agregata LBF-1-0016 Pseudomonas balearica LBF 1-0062 and Lysobacter concretionis LBF-1-0080) for oil degradation. AIP Conference Proceedings. 2018; 2024(1): 020043.
Zakaria MP, Bong CW, Vaezzadeh V. Fingerprinting of petroleum hydrocarbons in Malaysia using environmental forensic techniques: a 20-year field data review. Oil Spill Environ Forensics Case Stud. 2018;2018:345–72.
Acknowledgements
This study was sponsored by Geran Putra IPB (UPM Reference Code: UPM/700-2/1/GP-IPB/2013/9412400) and Geran Putra IPB (UPM Reference Code: UPM/700-2/1/GP/2018/9592200) awarded by Universiti Putra Malaysia (UPM). We also acknowledge the contributions of International Institute of Aquaculture and Aquatic Sciences/Department of Aquaculture, Universiti Putra Malaysia for laboratory equipment provided for conducting experiments.
Funding
Research on this publication was funded by: Geran Putra IPB (UPM Reference Code: UPM/700–2/1/GP-IPB/2013/9412400).
Author information
Authors and Affiliations
Contributions
We confirm that authors listed in this manuscript made substantial contributions to research design, data analysis and interpretation. They were also involved in drafting and critically revision of manuscripts’ intellectual content. They have given their final approval for publication.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
We the authors declare our consent for publication of original manuscript that has not been published before and is not currently being considered for publication elsewhere.
Competing interests
We wish to confirm that there are no known competing interests associated with this publication. We would like to draw the attention of the Editor to the following publication of one or more of us that refer to aspects of the manuscript presently being submitted: Isolation, identification and diesel-oil biodegradation capacities of indigenous hydrocarbon-degrading strains of Cellulosimicrobium cellulans and Acinetobacter baumannii from tarball at Terengganu beach, Malaysia: https://doi.org/10.1016/j.marpolbul.2016.03.060
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nkem, B.M., Halimoon, N., Yusoff, F.M. et al. Use of Taguchi design for optimization of diesel-oil biodegradation using consortium of Pseudomonas stutzeri, Cellulosimicrobium cellulans, Acinetobacter baumannii and Pseudomonas balearica isolated from tarball in Terengganu Beach, Malaysia. J Environ Health Sci Engineer 20, 729–747 (2022). https://doi.org/10.1007/s40201-022-00812-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40201-022-00812-3