Abstract
Background and Aims
Sulfonylureas are the most secondary prescribed oral anti-diabetic drug. Understanding its genetic role in pharmacodynamics can elucidate a considerable knowledge about personalized treatment in type 2 diabetes patients. This study aimed to assess the impact of KCNQ1 variants on sulfonylureas response among type 2 diabetes Iranian patients.
Methods and Results
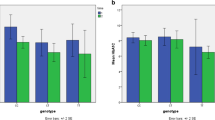
100 patients were recruited who were under sulfonylureas therapy for six months. 50 responder and 50 non-responder patients were selected. KCNQ1 variants were determined by the RFLP method, and their role in treatment response was assessed retrospectively. Patients with rs2237895 CC and AC genotypes demonstrated a significant decrement in FBS and HbA1c after treatment over patients with AA genotypes (All P < 0.001). Compared to the A allele, the odds ratio for treatment success between carriers with rs2237895 C allele was 4.22-fold (P < 0.001). Patients with rs2237892 CT heterozygous genotype exhibit a higher reduction rate in HbA1c and FBS than CC homozygotes (P=0.064 and P=0.079, respectively). The rs2237892 T allele carriers showed an odds ratio equals to 2.83-fold over C allele carriers in the responder group compared to the non-responder group (p=0.081).
Conclusion
Findings suggest that the KCNQ1 rs2237895 polymorphism is associated with the sulfonylureas response on Iranian type 2 diabetes patients.


Similar content being viewed by others
Abbreviations
- T2DM:
-
Type 2 diabetes mellitus
- T2D :
-
Type 2 diabetes
- SUR1 :
-
Sulfonylurea receptor 1
- SUR2A :
-
Sulfonylurea receptor 2A
- SUR2B :
-
Sulfonylurea receptor 2B
- RFLP :
-
Restriction fragment length polymorphism
- KCNQ1 :
-
Potassium Voltage-Gated Channel Subfamily Q Member 1
- SU :
-
Sulfonylurea
- BMI :
-
Body mass index
- FBS :
-
Fast blood sugar
- HbA1c :
-
Hemoglobin A1C
- HDL :
-
High-density lipid
- LDL :
-
Low-density lipid
- TC :
-
Total cholesterol
- TG :
-
Triglyceride
References
Cerf ME. Beta cell dysfunction and insulin resistance. Front Endocrinol. 2013;4:37.
Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442.
Esteghamati A, Larijani B, Aghajani MH, Ghaemi F, Kermanchi J, Shahrami A, et al. Diabetes in Iran: prospective analysis from first nationwide diabetes report of National Program for prevention and control of diabetes (NPPCD-2016). Sci Rep. 2017;7(1):1–10.
Pearson E, Liddell W, Shepherd M, Corrall R, Hattersley A. Sensitivity to sulphonylureas in patients with hepatocyte nuclear factor-1α gene mutations: evidence for pharmacogenetics in diabetes. Diabet Med. 2000;17(7):543–5.
Landman GW, de Bock GH, Van Hateren KJ, Van Dijk PR, Groenier KH, Gans RO, et al. Safety and efficacy of gliclazide as treatment for type 2 diabetes: a systematic review and meta-analysis of randomized trials. PLoS One. 2014;9(2):e82880.
Prescott L. Pathological and physiological factors affecting drug absorption, distribution, elimination, and response in man. Concepts in biochemical pharmacology: Springer. 1975:234–57.
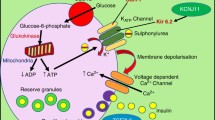
Gribble F, Ashcroft F. Differential sensitivity of beta-cell and extrapancreatic KATP channels to gliclazide. Diabetologia. 1999;42(7):845–8.
Proks P, Reimann F, Green N, Gribble F, Ashcroft F. Sulfonylurea stimulation of insulin secretion. Diabetes. 2002;51(suppl 3):S368–S76.
Eichelbaum M, Ingelman-Sundberg M, Evans WE. Pharmacogenomics and individualized drug therapy. Annu Rev Med. 2006;57:119–37.
Ventola CL. The role of pharmacogenomic biomarkers in predicting and improving drug response: part 2: challenges impeding clinical implementation. Pharmacy and therapeutics. 2013;38(10):624.
Loganadan N, Huri H, Vethakkan S, Hussein Z. Genetic markers predicting sulphonylurea treatment outcomes in type 2 diabetes patients: current evidence and challenges for clinical implementation. The pharmacogenomics journal. 2016;16(3):209–19.
Ordelheide A-M. Hrabě de Angelis M, Häring H-U, Staiger H. Pharmacogenetics of oral antidiabetic therapy. Pharmacogenomics. 2018;19(6):577–87.
Szabo M, Máté B, Csép K, Benedek T. Genetic approaches to the study of gene variants and their impact on the pathophysiology of type 2 diabetes. Biochem Genet. 2018;56(1):22–55.
Yazdi KV, Kalantar SM, Houshmand M, Rahmanian M, Manaviat MR, Jahani MR, et al. SLC30A8, CDKAL1, TCF7L2, KCNQ1 and IGF2BP2 are associated with type 2 diabetes mellitus in Iranian patients. Diabetes, Metabolic Syndrome and Obesity: Targets and Therapy. 2020;13:897.
Holmkvist J, Banasik K, Andersen G, Unoki H, Jensen TS, Pisinger C, et al. The type 2 diabetes associated minor allele of rs2237895 KCNQ1 associates with reduced insulin release following an oral glucose load. PLoS One. 2009;4(6):e5872.
Müssig K, Staiger H, Machicao F, Kirchhoff K, Guthoff M, Schäfer SA, et al. Association of type 2 diabetes candidate polymorphisms in KCNQ1 with incretin and insulin secretion. Diabetes. 2009;58(7):1715–20.
Yamagata K, Senokuchi T, Lu M, Takemoto M, Karim MF, Go C, et al. Voltage-gated K+ channel KCNQ1 regulates insulin secretion in MIN6 β-cell line. Biochem Biophys Res Commun. 2011;407(3):620–5.
Tan JT, Nurbaya S, Gardner D, Ye S, Tai ES, Ng DP. Genetic variation in KCNQ1 associates with fasting glucose andβ-cell function: a study of 3,734 subjects comprising three ethnicities living in Singapore. Diabetes. 2009;58(6):1445–9.
Jonsson A, Isomaa B, Tuomi T, Taneera J, Salehi A, Nilsson P, et al. A variant in the KCNQ1 gene predicts future type 2 diabetes and mediates impaired insulin secretion. Diabetes. 2009;58(10):2409–13.
Hu C, Wang C, Zhang R, Ma X, Wang J, Lu J, et al. Variations in KCNQ1 are associated with type 2 diabetes and beta cell function in a Chinese population. Diabetologia. 2009;52(7):1322–5.
Robbins J. KCNQ potassium channels: physiology, pathophysiology, and pharmacology. Pharmacol Ther. 2001;90(1):1–19.
Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40(9):1092.
Duan F, Guo Y, Zhang L, Chen P, Wang X, Liu Z, et al. Association of KCNQ1 polymorphisms with gliclazide efficacy in Chinese type 2 diabetic patients. Pharmacogenet Genomics. 2016;26(4):178–83.
Schroner Z, Dobrikova M, Klimcakova L, Javorsky M, Zidzik J, Kozarova M, et al. Variation in KCNQ1 is associated with therapeutic response to sulphonylureas. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research. 2011;17(7):CR392.
Yu W, Hu C, Zhang R, Wang C, Qin W, Lu J, et al. Effects of KCNQ1 polymorphisms on the therapeutic efficacy of oral antidiabetic drugs in Chinese patients with type 2 diabetes. Clinical Pharmacology & Therapeutics. 2011;89(3):437–42.
Li Q. Tang T-t, Jiang F, Zhang R, Chen M, yin J, et al. polymorphisms of the KCNQ1 gene are associated with the therapeutic responses of sulfonylureas in Chinese patients with type 2 diabetes. Acta Pharmacol Sin. 2017;38(1):80–9.
Alberti KGMM, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus. Provisional report of a WHO consultation. Diabet Med. 1998;15(7):539–53.
Drouin P, Standl E, Group DMS. Gliclazide modified release*: results of a 2-year study in patients with type 2 diabetes. Diabetes Obes Metab. 2004;6(6):414–21.
Roumie CL, Greevy RA, Grijalva CG, Hung AM, Liu X, Griffin MR. Diabetes treatment intensification and associated changes in HbA1c and body mass index: a cohort study. BMC Endocr Disord. 2016;16(1):1–9.
Schroner Z, Javorsky M, Tkacova R, Klimcakova L, Dobrikova M, Habalova V, et al. Effect of sulphonylurea treatment on glycaemic control is related to TCF7L2 genotype in patients with type 2 diabetes. Diabetes Obes Metab. 2011;13(1):89–91.
Schernthaner G, Grimaldi A, Di Mario U, Drzewoski J, Kempler P, Kvapil M, et al. GUIDE study: double-blind comparison of once-daily gliclazide MR and glimepiride in type 2 diabetic patients. Eur J Clin Investig. 2004;34(8):535–42.
Thulé PM, Umpierrez G. Sulfonylureas: a new look at old therapy. Current diabetes reports. 2014;14(4):473.
Kurukulasuriya LR, Sowers JR. Therapies for type 2 diabetes: lowering HbA1c and associated cardiovascular risk factors. Cardiovasc Diabetol. 2010;9(1):1–13.
MacDonald PE, Ha XF, Wang J, Smukler SR, Sun AM, Gaisano HY, et al. Members of the Kv1 and Kv2 voltage-dependent K+ channel families regulate insulin secretion. Mol Endocrinol. 2001;15(8):1423–35.
van Vliet-Ostaptchouk JV, van Haeften TW, Landman GW, Reiling E, Kleefstra N, Bilo HJ, et al. Common variants in the type 2 diabetes KCNQ1 gene are associated with impairments in insulin secretion during hyperglycaemic glucose clamp. PLoS One. 2012;7(3):e32148.
Acknowledgments
Hereby, we would like to thank the whole team of the Imam Khomeini hospital of Ahvaz and especially diabetes department nurses who have helped us greatly to conduct this study. Also, we want to declare our sincere gratitude to all participants who have aided us generously in this way.
Funding
This work was supported by Ahvaz Jundishapur University Medical Sciences (research project no: D-9712).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interest
We have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shakerian, S., Rashidi, H., Birgani, M.T. et al. KCNQ1 rs2237895 polymorphism is associated with the therapeutic response to sulfonylureas in Iranian type 2 diabetes mellitus patients. J Diabetes Metab Disord 21, 33–41 (2022). https://doi.org/10.1007/s40200-021-00931-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40200-021-00931-y