Abstract
Background
As a membrane G protein coupled receptors (GPCRs) family, gastrin/cholecystokinin-2 receptor (CCK2R) plays a key role in the initiation and development of gastric cancer.
Objectives
Targeting CCK2R by immunotherapeutics such as single-chain variable fragments (scFvs) may provide an effective treatment modality against gastric cancer. Thus, the main objective of this study was to isolate scFvs specific to CCK2R.
Methods
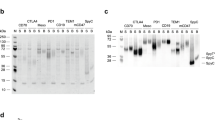
To isolate scFvs specific to the CCK2R, we capitalized on a semi-synthetic diverse phage antibody library (PAL) and a solution-phase biopanning process. The library was panned against a biotinylated peptide of the second extracellular loop (ECL2) of CCK2R. After four rounds of biopanning, the selected soluble scFv clones were screened by enzyme-linked immunosorbent assay (ELISA) and examined for specific binding to the peptide. The selected scFvs were purified using immobilized metal affinity chromatography (IMAC). The binding affinity and specificity of the scFvs were examined by the surface plasmon resonance (SPR), immunoblotting and flow cytometry assays and molecular docking using ZDOCK v3.0.2.
Results
Ten different scFvs were isolated, which displayed binding affinity ranging from 0.68 to 8.0 (nM). Immunoblotting and molecular docking analysis revealed that eight scFvs were able to detect the denatured form of CCK2R protein. Of the isolated scFvs, two scFvs showed high-binding affinity to the human gastric adenocarcinoma AGS cells.
Conclusions
Based on our findings, a couple of the selected scFvs showed markedly high-binding affinity to immobilized CCK2R peptide and CCK2R-overexpressing AGS cells. Therefore, these scFvs are proposed to serve as targeting and/or treatment agents in the diagnosis and immunotherapy of CCK2R-positive tumors.

ᅟ





Similar content being viewed by others
Abbreviations
- CCK2R:
-
Gastrin/cholecystokinin-2 receptor
- GPCRs:
-
G protein coupled receptors
- scFvs:
-
Single-chain variable fragments
- PAL:
-
Phage antibody library
- ECL2:
-
Extracellular loop
- ELISA:
-
Enzyme-linked immunosorbent assay
- IMAC:
-
Immobilized metal affinity chromatography
- SPR:
-
Surface plasmon resonance
- TM:
-
Transmembrane
- ICL:
-
Intracellular loops
- ECL:
-
Extracellular loops
- MALT:
-
Mucosa-associated lymphoid tissue lymphomas
- mAbs:
-
Monoclonal antibodies
- PAD:
-
Phage antibody display
- Ag:
-
Antigen
- g3p:
-
Gene 3 minor coat protein
- PDT:
-
Phage display technology
References
Matsueda S, Graham DY. Immunotherapy in gastric cancer. World J Gastroenterol. 2014;20(7):1657–66. https://doi.org/10.3748/wjg.v20.i7.1657.
Kawakami H, Okamoto I. MET-targeted therapy for gastric cancer: the importance of a biomarker-based strategy. Gastric cancer : official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association. 2016;19(3):687–95. https://doi.org/10.1007/s10120-015-0585-x.
Fahimi F, Tohidkia MR, Fouladi M, Aghabeygi R, Samadi N, Omidi Y. Pleiotropic cytotoxicity of VacA toxin in host cells and its impact on immunotherapy. Bioimpacts. 2017;7(1):59–71. https://doi.org/10.15171/bi.2017.08.
Fourmy D, Gigoux V, Reubi JC. Gastrin in gastrointestinal diseases. Gastroenterology. 2011;141(3):814–8 e1–3. https://doi.org/10.1053/j.gastro.2011.07.006.
Berna MJ, Tapia JA, Sancho V, Jensen RT. Progress in developing cholecystokinin (CCK)/gastrin receptor ligands that have therapeutic potential. Curr Opin Pharmacol. 2007;7(6):583–92. https://doi.org/10.1016/j.coph.2007.09.011.
Peeters MC, van Westen GJ, Li Q, AP IJ. Importance of the extracellular loops in G protein-coupled receptors for ligand recognition and receptor activation. Trends Pharmacol Sci. 2011;32(1):35–42. https://doi.org/10.1016/j.tips.2010.10.001.
Zhou JJ, Chen ML, Zhang QZ, Hu JK, Wang WL. Coexpression of cholecystokinin-B/gastrin receptor and gastrin gene in human gastric tissues and gastric cancer cell line. World J Gastroenterol. 2004;10(6):791–4.
McWilliams DF, Watson SA, Crosbee DM, Michaeli D, Seth R. Coexpression of gastrin and gastrin receptors (CCK-B and delta CCK-B) in gastrointestinal tumour cell lines. Gut. 1998;42(6):795–8.
Barderas R, Shochat S, Timmerman P, Hollestelle MJ, Martinez-Torrecuadrada JL, Hoppener JW, et al. Designing antibodies for the inhibition of gastrin activity in tumoral cell lines. Int J Cancer. 2008;122(10):2351–9. https://doi.org/10.1002/ijc.23395.
Demarest SJ, Hariharan K, Dong J. Emerging antibody combinations in oncology. MAbs. 2011;3(4):338–51.
Koohapitagtam M, Rungpragayphan S, Hongprayoon R, Kositratana W, Sirinarumitr T. Efficient amplification of light and heavy chain variable regions and construction of a non-immune phage scFv library. Mol Biol Rep. 2010;37(4):1677–83. https://doi.org/10.1007/s11033-009-9583-6.
Reichert JM. Metrics for antibody therapeutics development. MAbs. 2010;2(6):695–700.
Huang JX, Bishop-Hurley SL, Cooper MA. Development of anti-infectives using phage display: biological agents against bacteria, viruses, and parasites. Antimicrob Agents Chemother. 2012;56(9):4569–82. https://doi.org/10.1128/AAC.00567-12.
Pansri P, Jaruseranee N, Rangnoi K, Kristensen P, Yamabhai M. A compact phage display human scFv library for selection of antibodies to a wide variety of antigens. BMC Biotechnol. 2009;9:6. https://doi.org/10.1186/1472-6750-9-6.
Paschke M. Phage display systems and their applications. Appl Microbiol Biotechnol. 2006;70(1):2–11. https://doi.org/10.1007/s00253-005-0270-9.
Soltes G, Hust M, Ng KK, Bansal A, Field J, Stewart DI, et al. On the influence of vector design on antibody phage display. J Biotechnol. 2007;127(4):626–37. https://doi.org/10.1016/j.jbiotec.2006.08.015.
Whiteaker JR, Zhao L, Frisch C, Ylera F, Harth S, Knappik A, et al. High-affinity recombinant antibody fragments (Fabs) can be applied in peptide enrichment immuno-MRM assays. J Proteome Res. 2014;13(4):2187–96. https://doi.org/10.1021/pr4009404.
Noppe W, Plieva F, Galaev IY, Pottel H, Deckmyn H, Mattiasson B. Chromato-panning: an efficient new mode of identifying suitable ligands from phage display libraries. BMC Biotechnol. 2009;9:21. https://doi.org/10.1186/1472-6750-9-21.
Ahmad ZA, Yeap SK, Ali AM, Ho WY, Alitheen NB, Hamid M. scFv antibody: principles and clinical application. Clin Dev Immunol. 2012;2012:980250. https://doi.org/10.1155/2012/980250.
Cheng LS, Liu AP, Yang JH, Dong YQ, Li LW, Wang J, et al. Construction, expression and characterization of the engineered antibody against tumor surface antigen, p185(c-erbB-2). Cell Res. 2003;13(1):35–48. https://doi.org/10.1038/sj.cr.7290149.
Wang Y, Zhang X, Zhang C, Liu Y, Liu X. Isolation of single chain variable fragment (scFv) specific for Cry1C toxin from human single fold scFv libraries. Toxicon: Official Journal of the International Society on Toxinology. 2012;60(7):1290–7. https://doi.org/10.1016/j.toxicon.2012.08.014.
Huang L, Sato AK, Sachdeva M, Fleming T, Townsend S, Dransfield DT. Discovery of human antibodies against the C5aR target using phage display technology. J Mol Recognit: JMR. 2005;18(4):327–33. https://doi.org/10.1002/jmr.735.
Henderikx P, Kandilogiannaki M, Petrarca C. von Mensdorff-Pouilly S, Hilgers JH, Krambovitis E et al. human single-chain Fv antibodies to MUC1 core peptide selected from phage display libraries recognize unique epitopes and predominantly bind adenocarcinoma. Cancer Res. 1998;58(19):4324–32.
Hawkins RE, Russell SJ, Winter G. Selection of phage antibodies by binding affinity. Mimicking affinity maturation. J Mol Biol. 1992;226(3):889–96.
de Haard HJ, van Neer N, Reurs A, Hufton SE, Roovers RC, Henderikx P, et al. A large non-immunized human fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J Biol Chem. 1999;274(26):18218–30.
Zhang X, Liu Y, Zhang C, Wang Y, Xu C, Liu X. Rapid isolation of single-chain antibodies from a human synthetic phage display library for detection of bacillus thuringiensis (Bt) Cry1B toxin. Ecotoxicol Environ Saf. 2012;81:84–90. https://doi.org/10.1016/j.ecoenv.2012.04.021.
Retter I, Althaus HH, Munch R, Muller W. VBASE2, an integrative V gene database. Nucleic Acids Res. 2005;33(Database issue):D671–4. https://doi.org/10.1093/nar/gki088.
Eteshola E. Isolation of scFv fragments specific for monokine induced by interferon-gamma (MIG) using phage display. J Immunol Methods. 2010;358(1–2):104–10. https://doi.org/10.1016/j.jim.2010.04.003.
Johns M, George AJ, Ritter MA. In vivo selection of sFv from phage display libraries. J Immunol Methods. 2000;239(1–2):137–51.
Wu S, Ke A, Doudna JA. A fast and efficient procedure to produce scFvs specific for large macromolecular complexes. J Immunol Methods. 2007;318(1–2):95–101. https://doi.org/10.1016/j.jim.2006.10.005.
Sushma K, Vijayalakshmi MA, Krishnan V, Satheeshkumar PK. Cloning, expression, purification and characterization of a single chain variable fragment specific to tumor necrosis factor alpha in Escherichia coli. J Biotechnol. 2010;156(4):238–44. https://doi.org/10.1016/j.jbiotec.2011.06.039.
Hsu FY, Chou LF, Hor LI, Chang HY. A human single-chain variable fragment targeting to Vibrio vulnificus RtxA toxin. J Microbiol Methods. 2011;84(1):94–100. https://doi.org/10.1016/j.mimet.2010.11.006.
Poul MA, Becerril B, Nielsen UB, Morisson P, Marks JD. Selection of tumor-specific internalizing human antibodies from phage libraries. J Mol Biol. 2000;301(5):1149–61. https://doi.org/10.1006/jmbi.2000.4026.
Martinez-Torrecuadrada J, Cifuentes G, Lopez-Serra P, Saenz P, Martinez A, Casal JI. Targeting the extracellular domain of fibroblast growth factor receptor 3 with human single-chain Fv antibodies inhibits bladder carcinoma cell line proliferation. Clin Cancer Res. 2005;11(17):6280–90. https://doi.org/10.1158/1078-0432.CCR-05-0282.
Kim HY, Wang X, Wahlberg B, Edwards WB. Discovery of hapten-specific scFv from a phage display library and applications for HER2-positive tumor imaging. Bioconjug Chem. 2014;25(7):1311–22. https://doi.org/10.1021/bc500173f.
Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2017;1654:39–54. https://doi.org/10.1007/978-1-4939-7231-9_4.
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–12. https://doi.org/10.1002/jcc.20084.
Johansson MU, Zoete V, Michielin O, Guex N. Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinformatics. 2012;13:173. https://doi.org/10.1186/1471-2105-13-173.
Pourseif MM, Moghaddam G, Daghighkia H, Nematollahi A. Omidi Y. a novel B- and helper T-cell epitopes-based prophylactic vaccine against Echinococcus granulosus. Bioimpacts. 2018;8(1):39–52. https://doi.org/10.15171/bi.2018.06.
Dunbrack RL Jr. Rotamer libraries in the 21st century. Curr Opin Struct Biol. 2002;12(4):431–40.
Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Web Server):W407–10. https://doi.org/10.1093/nar/gkm290.
Eisenberg D, Luthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404.
Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–9. https://doi.org/10.1002/pro.5560020916.
Pourseif MM, Moghaddam G, Naghili B, Saeedi N, Parvizpour S, Nematollahi A, et al. A novel in silico minigene vaccine based on CD4(+) T-helper and B-cell epitopes of EG95 isolates for vaccination against cystic echinococcosis. Comput Biol Chem. 2018;72:150–63. https://doi.org/10.1016/j.compbiolchem.2017.11.008.
Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG, et al. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50(3):437–50. https://doi.org/10.1002/prot.10286.
Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52(1):80–7. https://doi.org/10.1002/prot.10389.
Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One. 2011;6(9):e24657. https://doi.org/10.1371/journal.pone.0024657.
Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8(2):127–34.
Reichert JM. Antibodies to watch in 2017. MAbs. 2017;9(2):167–81. https://doi.org/10.1080/19420862.2016.1269580.
Majidi J, Barar J, Baradaran B, Abdolalizadeh J, Omidi Y. Target therapy of cancer: implementation of monoclonal antibodies and nanobodies. Hum Antibodies. 2009;18(3):81–100. https://doi.org/10.3233/hab-2009-0204.
Mondon P, Dubreuil O, Bouayadi K, Kharrat H. Human antibody libraries: a race to engineer and explore a larger diversity. Front Biosci. 2008;13:1117–29.
Sergeeva A, Kolonin MG, Molldrem JJ, Pasqualini R, Arap W. Display technologies: application for the discovery of drug and gene delivery agents. Adv Drug Deliv Rev. 2006;58(15):1622–54. https://doi.org/10.1016/j.addr.2006.09.018.
Ebrahimizadeh W, Rajabibazl M. Bacteriophage vehicles for phage display: biology, mechanism, and application. Curr Microbiol. 2014;69(2):109–20. https://doi.org/10.1007/s00284-014-0557-0.
Zhao A, Tohidkia MR, Siegel DL, Coukos G, Omidi Y. Phage antibody display libraries: a powerful antibody discovery platform for immunotherapy. Crit Rev Biotechnol. 2016;36(2):276–89. https://doi.org/10.3109/07388551.2014.958978.
Khajeh S, Tohidkia MR, Aghanejad A, Mehdipour T, Fathi F, Omidi Y. Phage display selection of fully human antibody fragments to inhibit growth-promoting effects of glycine-extended gastrin 17 on human colorectal cancer cells. Artif Cells Nanomed Biotechnol. 2018:1–9. https://doi.org/10.1080/21691401.2018.1478846.
Fahimi F, Sarhaddi S, Fouladi M, Samadi N, Sadeghi J, Golchin A, et al. Phage display-derived antibody fragments against conserved regions of VacA toxin of helicobacter pylori. Appl Microbiol Biotechnol. 2018;102:6899–913. https://doi.org/10.1007/s00253-018-9068-4.
Tohidkia MR, Sepehri M, Khajeh S, Barar J, Omidi Y. Improved soluble ScFv ELISA screening approach for antibody discovery using phage display technology. SLAS Discov. 2017;22(8):1026–34. https://doi.org/10.1177/2472555217701059.
Tohidkia MR, Asadi F, Barar J, Omidi Y. Selection of potential therapeutic human single-chain Fv antibodies against cholecystokinin-B/gastrin receptor by phage display technology. BioDrugs : Clinical Immunotherapeutics, Biopharmaceuticals and Gene Therapy. 2013;27(1):55–67. https://doi.org/10.1007/s40259-012-0007-0.
Abdolalizadeh J, Nouri M, Zolbanin JM, Barzegari A, Baradaran B, Barar J, et al. Targeting cytokines: production and characterization of anti-TNF-alpha scFvs by phage display technology. Curr Pharm Des. 2013;19(15):2839–47.
Tohidkia MR, Barar J, Asadi F, Omidi Y. Molecular considerations for development of phage antibody libraries. J Drug Target. 2012;20(3):195–208. https://doi.org/10.3109/1061186X.2011.611517.
Abdolalizadeh J, Nouri M, Zolbanin JM, Baradaran B, Barzegari A, Omidi Y. Downstream characterization of anti-TNF-alpha single chain variable fragment antibodies. Hum Antibodies. 2012;21(1–2):41–8. https://doi.org/10.3233/HAB-2012-0260.
Hutchings CJ, Koglin M, Marshall FH. Therapeutic antibodies directed at G protein-coupled receptors. MAbs. 2010;2(6):594–606. https://doi.org/10.4161/mabs.2.6.13420.
Kmiecik S, Jamroz M, Kolinski M. Structure prediction of the second extracellular loop in G-protein-coupled receptors. Biophys J. 2014;106(11):2408–16. https://doi.org/10.1016/j.bpj.2014.04.022.
Silacci M, Brack S, Schirru G, Marlind J, Ettorre A, Merlo A, et al. Design, construction, and characterization of a large synthetic human antibody phage display library. Proteomics. 2005;5(9):2340–50. https://doi.org/10.1002/pmic.200401273.
Lobova D, Cizek A, Celer V. The selection of single-chain Fv antibody fragments specific to Bhlp 29.7 protein of Brachyspira hyodysenteriae. Folia Microbiol. 2008;53(6):517–20. https://doi.org/10.1007/s12223-008-0081-3.
Villa A, Lovato V, Bujak E, Wulhfard S, Pasche N, Neri D. A novel synthetic naive human antibody library allows the isolation of antibodies against a new epitope of oncofetal fibronectin. MAbs. 2011;3(3):264–72.
Zhang Y, Pool C, Sadler K, Yan HP, Edl J, Wang X, et al. Selection of active ScFv to G-protein-coupled receptor CCR5 using surface antigen-mimicking peptides. Biochemistry. 2004;43(39):12575–84. https://doi.org/10.1021/bi0492152.
Gupta A, Decaillot FM, Gomes I, Tkalych O, Heimann AS, Ferro ES, et al. Conformation state-sensitive antibodies to G-protein-coupled receptors. J Biol Chem. 2007;282(8):5116–24. https://doi.org/10.1074/jbc.M609254200.
Koide A, Koide S. Monobodies: antibody mimics based on the scaffold of the fibronectin type III domain. Methods Mol Biol. 2007;352:95–109. https://doi.org/10.1385/1-59745-187-8:95.
Vazquez-Lombardi R, Phan TG, Zimmermann C, Lowe D, Jermutus L, Christ D. Challenges and opportunities for non-antibody scaffold drugs. Drug Discov Today. 2015;20(10):1271–83. https://doi.org/10.1016/j.drudis.2015.09.004.
Moghimi SM, Rahbarizadeh F, Ahmadvand D, Parhamifar L. Heavy chain only antibodies: a new paradigm in personalized HER2+ breast Cancer therapy. BioImpacts : BI. 2013;3(1):1–4. https://doi.org/10.5681/bi.2013.009.
Liebman MA, Williams BR, Daley KM, Sharon J. Generation and preliminary characterization of an antibody library with preferential reactivity to human colorectal cancer cells as compared to normal human blood cells. Immunol Lett. 2004;91(2–3):179–88. https://doi.org/10.1016/j.imlet.2003.12.002.
Acknowledgments
The authors are grateful to the technical support provided by the Research Center for Pharmaceutical Nanotechnology (RCPN) at Tabriz University of Medical Sciences.
Funding
The authors are grateful for the financial support provided by the Tabriz University of Medical Sciences (grant# 93003 and 93010).
Author information
Authors and Affiliations
Contributions
SJR and MM contributed to the data analysis and interpretation, and MMP was involved in the bioinformatics analysis, YP checked the grammar, MRT, and YO designed and supervised the study, conducted data analysis and interpretation, SJR, JB, MMP, MRT and YO wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors of this study declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jalilzadeh-Razin, S., Mantegi, M., Tohidkia, M.R. et al. Phage antibody library screening for the selection of novel high-affinity human single-chain variable fragment against gastrin receptor: an in silico and in vitro study. DARU J Pharm Sci 27, 21–34 (2019). https://doi.org/10.1007/s40199-018-0233-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40199-018-0233-1