Abstract
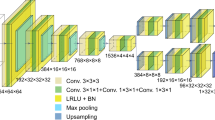
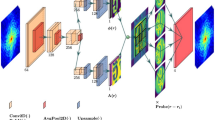
An important leap forward for the 3D community is the possibility to perform non-destructive 3D microstructural imaging in the home laboratories. This possibility is profiled by a recently developed technique—laboratory X-ray diffraction contrast tomography (LabDCT). As diffraction spots in LabDCT images are the basis for 3D reconstruction of microstructures, it is critical to get their identification as precise as possible. In the present work we use a deep learning (DL) routine to optimize the identification of the spots. It is shown that by adding an artificial simple constant background noise to a series of forward simulated LabDCT diffraction images, DL can be trained and then learn to remove high frequency noise and low frequency radial gradients in brightness in the real experimental LabDCT images. The training of the DL routine is unsupervised in the sense that no human intervention is needed for labelling the data. The reduction in high frequency noise and low frequency radial gradients in brightness is demonstrated by comparing line profile scans through the experimental and the DL output images. Finally, the implications of this reduction procedure on the spot identification are analysed and possible improvements are discussed.





Similar content being viewed by others
References
Poulsen H, Fæster S, Lauridsen E, Schmidt S, Suter R, Lienert U, Margulies L, Lorentzen T, Juul Jensen D (2001) Three-dimensional maps of grain boundaries and the stress state of individual grains in polycrystals and powders. J. Appl. Crystallogr. 34:751–756
Tischler J, Liu W, Xu R (2019) Micro-diffraction in 3D. In: Proceedings of 40th Risø international symposium on materials science: metal microstructures in 2D, 3D and 4D, pp 329–334
Zhang, Y (2019) Quantification of local boundary migration in 2D/3D. In: IOP conference series: materials science and engineering, vol 580, 012015
Bhattacharya A, Shen YF, Hefferan CM, Li SF, Lind J, Suter RM, Rohrer GS (2019) Three-dimensional observations of grain volume changes during annealing of polycrystalline Ni. Acta Mater 167:40–50
Hefferan CM, Lind J, Li SF, Lienert U, Rollett AD, Suter RM (2012) Observation of recovery and recrystallization in high-purity aluminum measured with forward modeling analysis of high-energy diffraction microscopy. Acta Mater 60(10):4311–4318
Bachmann F, Bale H, Gueninchault N, Holzner C, Lauridsen EM (2019) 3D grain reconstruction from laboratory diffraction contrast tomography. J Appl Crystallogr 52(3):643–651
Oddershede J, Sun J, Gueninchault N, Bachmann F, Bale H, Holzner C, Lauridsen E (2019) Non-destructive characterization of polycrystalline materials in 3d by laboratory diffraction contrast tomography. Integr Mater Manuf Innov 8(2):217–225
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. https://arxiv.org/abs/1505.04597
Johnson J, Alahi A, Li FF (2016) Perceptual losses for real-time style transfer and super-resolution. https://arxiv.org/abs/1603.08155
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. https://arxiv.org/abs/1409.1556
Fang H, Juul Jensen D, Zhang Y (2019) An efficient method to improve the spatial resolution of laboratory x-ray diffraction contrast tomography. In: IOP conference series: materials science and engineering, vol 580 (1) 012030
Boone JM, Seibert JA (1997) An accurate method for computer-generating tungsten anode x-ray spectra from 30 to 140 kv. Med Phys 24(11):1661–1670
Fang H, Juul Jensen D, Zhang Y (2020) A flexible and standalone forward simulation model for laboratory X-ray diffraction contrast tomography. Acta Crystallogr. https://doi.org/10.1107/S2053273320010852
https://github.com/fastai/course-v3/blob/master/nbs/dl1/lesson7-superres.ipynb
Acknowledgements
This work is financially supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (M4D—Grant Agreement No. 788567).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hovad, E., Fang, H., Zhang, Y. et al. Unsupervised Deep Learning for Laboratory-Based Diffraction Contrast Tomography. Integr Mater Manuf Innov 9, 315–321 (2020). https://doi.org/10.1007/s40192-020-00189-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40192-020-00189-x