Abstract
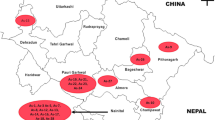
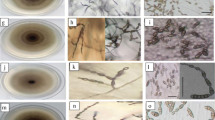
Early blight (EB), caused by the fungus, Alternaria solani, is one of the most destructive diseases of tomatoes and other solanaceous crops; particularly in warm and humid climate. This study was targeted to explore the genetic and pathogenic diversity of A. solani from major tomato producing states of India. Thirty-three isolates were chosen for this study. These isolates exhibited considerable intra as well as inter-state variation. The phylogenetic tree generated with the ISSR sequences confirmed this result. Aggressiveness of the isolates towards susceptible tomato genotype was assessed in vitro, using detached leaf method. Considerable amount of variability in virulence was observed among the isolates. Specific activity of polygalacturonase and pectin methyl esterase was also estimated, to observe the relation among these, with the virulence of isolates. The information generated in the present study provides initial data on the population variability of the EB pathogen. It could be a valuable aid for tomato breeding strategies, aimed at obtaining cultivars with resilient resistance. This will provide a basis for planning disease protection strategies for sustainable agriculture which is required for producing crop plants which harmonize with the environment.





Similar content being viewed by others
References
Gwary DM, Nahunnaro H (1998) Epiphytotics of early blight of tomatoes in northeastern Nigeria. Crop Prot 17:619–624
Milgroom MG, Peever TL (2003) Population biology of plant pathogens: the synthesis of plant disease epidemiology and population genetics. Plant Dis 87:608–617
Bateman DF, Basham HG (1976) Degradation of plant cell walls and membranes by microbial enzymes. In: Heitefuss R, Williams PH (eds) Physiological plant pathology. Springer, Berlin, pp 316–355
Walton JD (1994) Deconstructing the cell wall. Plant Physiol 104:1113–1118
Holtz G, Knox-Davies PS (1985) Production of pectic enzymes by Fusarium oxysporum f. sp. cepae and its involvement in onion bulb rot. J Phytopathol 112:69–80
Baayen RP, van Dreven F, Krijger MC, Wallwijk C (1997) Genetic diversity in Fusarium oxysporum f. sp. dianthi and Fusarium redolens f. sp. dianthi. Eur J Plant Pathol 103:395–408
Lathar PK, Sharma A, Thakur I (2010) Isolation and random amplified polymorphic DNA (RAPD) analysis of wild yeast species from 17 different fruits. J Yeast Fungal Res 18:146–151
Fegan M, Manners JM, Maclean DJ, Irwin JA, Samuels KD, Holdom DG, Li DP (1993) Random amplified polymorphic DNA markers reveal a high degree of genetic diversity in the entomopathogenic fungus Metarhizium anisopliae var. anisopliae. J Gen Microbiol 139:2075–2081
Kim Y, Choi SJ, Choi C (2017) An Efficient PCR-RFLP Method for the rapid identification of Korean Pyropia Species. Molecules. https://doi.org/10.3390/molecules22122182
Nath VS, Senthil M, Hegde VM, Jeeva ML, Misra RS, Veena SS, Raj M (2013) Genetic diversity of Phytophthora colocasiae isolates in India based on AFLP analysis. 3. Biotechnology 3(4):297–305. https://doi.org/10.1007/s13205-012-0101-55
Velez P, Quintero CA, Merino G, Gasca-Pineda J, González MC (2016) An ISSR-based approach to assess genetic diversity in the marine arenicolous fungus Corollospora maritima sensu lato. Mycoscience 57(3):187–195
Del-Prado R, Cubas P, Lumbsch HT, Divakar PK, Blanco O, de Paz GA, Molina MC, Crespo A (2010) Genetic distances within and among species in monophyletic lineages of Parmeliaceae (Ascomycota) as a tool for taxon delimitation. Mol Phylogenet Evol 56(1):125–133
Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A (2000) Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol 50(3):1351–1371
Vialle A, Feau N, Allaire M, Didukh M, Martin F, Moncalvo JM, Hamelin RC (2009) Evaluation of mitochondrial genes as DNA barcode for Basidiomycota. Mol Ecol Resour 9:99–113
Bagherabadi S, Zafari D, Soleimani MJ (2015) Genetic diversity of Alternaria alternata isolates causing potato brown leaf spot using ISSR markers in Iran. J Plant Pathol Microb 6:286. https://doi.org/10.4172/2157-7471.1000286
Troncoso-Rojas R, Báez-Flores María Elena, Pryor Barry, Garcíaand Hugo S, Tiznado-Hernández Martín-Ernesto (2013) Inter simple sequence repeat polymorphism in Alternaria genomic DNA exposed to lethal concentrations of Isothiocyanates. Afr J Microbil Res 7(10):838–852
Upadhyay P, Singh PC, Sinha B, Singh M, Kumar R (2009) Sources of resistance against early blight (Alternaria solani) in tomato (Solanum lycopersicum). Ind J Agric Sci 79:752–753
Weber B, Halterman DA (2012) Analysis of genetic and pathogenic variation of Alternaria solani from a potato production region. Eur J Plant Pathol 134:847–858
Kumari S, Tayal P, Sharma E, Kapoor R (2014) Analyses of genetic and pathogenic variability among Botrytis cinerea isolates Microbio Res 169(11):862–872
Balaban MO, Arreola AG, Marshall M, Peplow A, Wei CI, Cornel J (1991) Inactivation of pectinesterase in orange juice by supercritical carbon dioxide. J Food Sci 56:743–746
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Casadevall A, Pirofski L-A (1999) Host–pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect Immun 67:3703–3713
Cervone F, De Lorenzo G, Salvi G, Camardella L (1986). Molecular evolution of fungal polygalacturonase. In: Bailey J (ed) Biology and molecular biology of plant–pathogen interactions. NATOASI series, vol H1. Springer, Berlin, pp 385–392
Brett C, Waldron K (1990) Physiology and biochemistry of plant cell walls. In: Black M, Chapman J (eds) Topics in plant physiology. Unwin Hyman, London, pp 6–57
de Vries RP, Visser J (2001) Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol Mol Biol Rev 65:497–522
Martinez SP, Snowdon R, Pons-Kuhnemann J (2004) Variability of Cuban and international populations of Alternaria solani from different hosts and localities: AFLP genetic analysis. Eur J Plant Pathol 110(4):399–409
O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci USA 95:2044–2449
Prins TW, Tudzynski P, Von Tiedemann A, Tudzynski B, ten Have A, Hansen ME (2000) Infection strategies of Botrytis cinerea and related necrotrophic pathogens. In: Kronstad J (ed) Fungal pathology. Kluwer, Dordrecht, pp 33–64. https://doi.org/10.1007/978-94-015-9546-9_2
Gui Z, Gao J, Xin N, Wang Y, Yongshuo P, Liu H, Yuan Q, Li X (2016) Association of polygalacturonase-inhibiting protein gene 2 (MsPGIP2) to common leaf spot resistance in alfalfa. Eur J Plant Pathol 144:245–256
An SH, Sohn KH, Choi HW, Hwang IS, Lee SC, Hwang BK (2008) Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228(1):61–78
Acknowledgements
The authors are thankful to University Grant Commission, New Delhi, India for providing Post-Doctoral [Fellowship F.15-1/2013-14/PDFWM-2013-14-GE-UTT-20193 (SA-II)] and Department of Botany, University of Delhi, Delhi for providing laboratory facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest in this publication.
Additional information
Significance statement
Diversity assessment among A. solani isolates from different states of India will provide valuable aid for resistance breeding strategies and basis for planning disease protection for sustainable agriculture.
Rights and permissions
About this article
Cite this article
Upadhyay, P., Ganaie, S.H. & Singh, N. Diversity Assessment Among Alternaria solani Isolates Causing Early Blight of Tomato in India. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 89, 987–997 (2019). https://doi.org/10.1007/s40011-018-1017-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-018-1017-6