Abstract
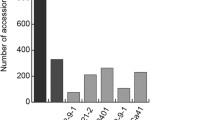
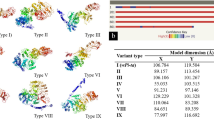
Rice blast is one of the important fungal diseases of rice, caused by Magnaporthe oryzae. Rice blast can be effectively managed by the deployment of R-genes. Pi2 (Piz-5) is one of the major blast resistant genes effective against pathogen populations in various parts of India. In the present study, Pi2 gene was mined from 83 native landraces of rice from Karnataka state, India which revealed the presence of Pi2 in 60 landraces with the frequency of 72.28 %. Further, by using sequencing based allele mining approach, allelic variants of Pi2 from 16 landraces were analysed. High nucleotide variations (InDels) in the middle region of the conserved domain sequence of Pi2 in all variants were obtained. 469 polymorphic sites were observed with a total 570 mutations and 191 InDels among 16 Pi2 variants. Among Pi2 resistant and susceptible variants, 206 mutations with 29 InDel events were observed. Comparative modeling and Ramachandran plot of Pi2 protein variants from resistant and susceptible landraces were studied which revealed the significant differences among them. One potential novel Pi2 allelic variant from landrace Vanasurya was identified for the first time. This novel effective allelic variant can be used in rice breeding programmes to enhance blast resistance.






Similar content being viewed by others
References
Khush GS (2005) What it will take to feed 5.0 billion rice consumers in 2030. Plant Molol Biol 59:1–6
Kumar A, Kumar S, Kumar R, Kumar V, Prasad V, Kumar N, Singh D (2010) Identification of blast resistance expression in rice genotypes using molecular markers (RAPD and SCAR). Afr J Biotechnol 9(24):3501–3509
Rola AC and Widawsky DA (1998) Impact of rice research. In: Proceedings of international conference on impact of rice research. TDRI, IRRI, Phillipines, pp 135–158
Couch BC, Kohn LM (2002) A multilocus gene genealogy concordant with host preference indicates segregation of a new species Magnaporthe oryzae from M. grisea. Mycologia 94:683–693
Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu JR, Pan H, Read ND, Lee YH, Carbone I, Brown D, Oh YY, Donofrio N, Jeong JS, Soanes DM, Djonovic S, Kolomiets E, Rehmeyer C, Li W, Harding M, Kim S, Lebrun MH, Bohnert H, Coughlan S, Butler J, Calvo S, Ma LJ, Nicol R, Purcell S, Nusbaum C, Galagan JE, Birren BW (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434(7036):980–986
Talbot NJ (2003) On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu Rev Microbiol 57:177–202
Sharma TR, Rai AK, Gupta SK, Vijayan J, Devanna BN, Ray S (2012) Rice blast management through host plant resistance: retrospect and prospects. Agric Res 1:37–52
Das A, Soubam D, Singh PK, Thakur S, Singh NK, Sharma TR (2012) A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct Integr Genom 12:215–228
Chen DH, Zeigler RS, Ahn SW, Nelson RJ (1996) Phenotypic characterization of the rice blast resistance gene Pi2(t). Plant Dis 80:52–56
Girish Kumar K, Hittalmani S, Srinivasachary, Shashidharhe (2000) Marker assisted backcross gene introgression of major genes for blast resistance in rice. Adv Rice Blast Res 15:43–53
Lavanya B, Gnanamanickam SS (2000) Molecular tools for characterization of rice blast pathogen (Magnaporthe grisea) population and molecular marker-assisted breeding for disease resistance. Curr Sci 78:248–257
Barclay A (2004) Feral play: crop scientist use wild crosses to breed into cultivated rice varieties the hardiness of their wild kin. Rice Today 3(1):14–19
Khush GS and Jena KK (2009) Current status and future prospects for research on blast resistance in rice (Oryza sativa L.). In: Wang GL, Valent B (eds) Advances in genetics, genomics and control of rice blast disease. Springer, Netherlands, pp 1–10
Ram Kumar G, Sakthivel K, Sundaram RM, Neeraja CN, Balachandran SM, Shobha Rani N, Viraktamath BC, Madhav MS (2010) Allele mining in crops: prospects and potentials. J Biotechnol Adv 28:451–461
Vanaja T, Singh R, Randhawa GJ (2010) Genetic relationships among a collection of Indica rice (Oryza sativa) genotypes of Kerala revealed by SSR markers. Indian J Agric Sci 80(3):191–197
Prashanthi SK, Hanamaratti NG, Salunke P (2009) A search for blast resistance sources from traditional rice collection of Karnataka. In: 5th international conference on plant pathology in the globalized era, New Delhi, p 362
Murray HG, Thompson WF (1980) Rapid isolation of high molecular weight DNA. Nucleic Acids Res 8:4321–4325
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likely-hood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ (eds) Evolving genes and proteins. Academic Press, New York, pp 97–166
Bhat RS, Gowda MVC (2004) Speciality native rice (Oryza sativa L.) germplasm of Uttara Kannada, India. Plant Genet Resour Newsl 140:42–47
Hanamaratti NG, Prashanthi SK, Salimath PM, Hanchinal RR, Mohankumar HD, Parameshwarappa KG, Raikar SD (2008) Traditional landraces of rice in Karnataka: reservoirs of valuable traits. Curr Sci 94(2):242–247
Hittalmani S, Kahani F, Dhanagond SM, Mohan Rao A (2013) DNA marker characterization for allele mining of blast and bacterial leaf blight resistant genes and evaluation for grain yield. Afr J Biotechnol 12(18):2331–2340
Kiyosawa S (1982) Genetic and epidemical modeling of breakdown of plant disease resistance. Annu Rev Phytopathol 20:93–117
Lattterell FM, Rossi AE (1986) Longevity and pathogenic stability of Pyricularia oryzae. Phytopathology 76:231–235
Latha R, Rubia L, Bennett J, Swaminathan MS (2004) Allele mining for stress tolerance genes in Oryza species and related germplasm. Mol Biotechnol 27:101–108
Thakur S, Gupta YK, Singh PK, Rathour R, Variar M, Prashanthi SK, Singh AK, Singh UD, Chand D, Rana JC, Singh NK, Sharma TR (2013) Molecular diversity in rice blast resistance gene Pi-ta makes it highly effective against dynamic population of Magnaporthe oryzae. Funct Integr Genom. doi:10.1007/s10142-013-0325-4
Thakur S, Singh PK, Rathour R, Variar M, Prashanthi SK, Singh AK, Singh UD, Chand D, Chand D, Singh NK, Sharma TR (2012) Positive selection pressure on rice resistance allele Piz-t makes it divergent in Indian landraces. J Plant Interact 8:34–44
Huynen MA, Bork P (1998) Measuring genome evolution. Proc Natl Acad Sci USA 95:5849–5856
Takken FL, Tameling WI (2009) To nibble at plant resistance proteins. Science 324:744–746
Lovell SC, Davis IW, Arendall WB, de Bakker P, Word JM, Prisant MG, Richardson JS, Richardson DC (2002) Structure validation by C-alpha geometry: phi, psi and C-beta deviation. Proteins Struct Funct Genet 50:437–450
Zhou B, Qu S, Liu G, Dolan M, Sakai H, Lu G, Bellizzi M, Wang GL (2006) The eight amino acid differences within three leucine rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol Plant Microbe Interact 19(11):1216–1228
Tekaia F (1999) The genomic tree as revealed from whole proteome comparisons. Genome Res 9:550–557
Acknowledgments
The first author is thankful to Department of Biotechnology (DBT), Govt. of India for providing fellowship during his master’s program. Authors are also thankful to NCBI and various authors for making available sequence data of reference Pi2 in the public domain.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ingole, K.D., Prashanthi, S.K. & Krishnaraj, P.U. Mining of Blast Resistance Gene, Pi2 and Its Novel Allelic Variant from Landraces of Rice from Karnataka. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 87, 1429–1441 (2017). https://doi.org/10.1007/s40011-016-0715-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-016-0715-1