Abstract
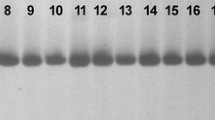
Tossa jute (Corchorus olitorius L.) is an important lingo-cellulosic bast fibre-crop. It provides biodegradable and environment friendly fibre next to cotton, in terms of usage, global consumption, production, and availability. Narrow genetic diversity of the crop is the major hurdle, which is a demand at priority for any crop improvement programme. In the current investigation 138 jute genotypes of C. olitorius were characterized with ten jute specific SSR markers. A total of 23 alleles were amplified with an average of 2.3 alleles per locus and the PIC value ranged from 0.13 to 0.76 with an average of 0.455. The un-weighted pair-group method with arithmetic average cluster analysis of the 138 jute genotypes depicted a dendrogram using DARWIN, which divided the genetic resource into three major clusters. The study indicated the utility of SSR primers for providing useful and high levels of polymorphism for individual plant genotypes even with a narrow genetic base. Based on cluster analysis the most divergent genotypes identified were OIJ 167 (from Indonesia), OIM 058 and OIM 059 (India), however based on the agronomic traits as maximum plant height, basal diameter and fibre weight they were OIJ 245, OIJ (153 and 161) and OIJ 040, respectively.



Similar content being viewed by others
Notes
References
Kundu BC (1951) Origin of jute. Indian J. Genetics 2:95–99
Kirby RH (1963) Vegetable fibres. Interscience Publishers, New York
Alam SS, Rahman ANMRB (2000) Karyotype analysis of three Corchorus species. Cytologia 65:443–446
Singh DP (1976) Jute: Evolution of crop plants. In: Simonds NW (ed) Longman Publishers Company, London, pp 290–291
Mondal R (2000) Diversified jute products. International Jute Organization Bangladesh, Dhaka, pp 115–118
Purseglove JW (1968) Tropical crops-dicotyledons, 2. Longman, Green and Co., Ltd., London
Alverson WS, Whitlock BA, Nyffeler R, Bayer C, Baum DA (1999) Phylogeny of the core Malvales: evidence from ndhF sequence data. Am J Bot 86:1474–1486
Whitlock BA, Karol KG, Alverson WS (2003) Chloroplast DNA sequences confirm the placement of the enigmatic Oceanopapaver within Corchorus (Grewioideae: Malvaceae s. l., formerly Tiliaceae). Int J Plant Sci 164:35–41
Heywood VH, Brummitt RK, Culham A, Seberg O (2007) Flowering plant families of the world. R Bot Gard, Kew
Kundu BC (1956) Jute world’s foremost bast fibre. Botany, agronomy, disease and pests. Eco Bot 10:103–133
Saunders M (2006) Recovery plan for the endangered native jute species, Corchoruous cunninghamee F. Muell in Queensland (2001–06). Natural Heritage Trust, Canberra, pp 1–29
Mahapatra AK, Saha A (2008) Genetic resources: jute and allied fibre corps. Jute and allied fibre updates. Central Research Institute for Jute and Allied Fibres, Kolkata, pp 18–37
Benor S, Blattner FR, Demissew S, Hammer K (2010) Collection and ethnobotanical investigation of Corchorus species in Ethiopia: potential leafy vegetables for dry regions. Genetic Res Crop Evol 57:293–306
Edmonds JM (1990) Herbarium survey of African Corchorus L. species. Systematic and ecogeographic studies on crop gene pools 4. International Board for Plant Genetic Resources, Rome
Ghosh T (1983) Handbook on jute. FAO, Rome, p 219
Bhaduri PN, Bairagi P (1968) Interspecific hybridization in jute (Corchorus capsularis x C. olitorius). Sci Cult 34:355–357
La Farge T, Friedman ST, Cock CG (1997) Improvement of fiber crops using genetics and biotechnology. In: Powell RM et al (eds) Paper and composites from agro-based resources. CRC Lewis Publishers, Boca Raton, pp 39–59
Anonymous (1997) General recommendation. In: Proceedings of the first workshop on jute DUS testing, organized by Strengthening Seed Certification Agency Project, Seed Certification Agency, Gazipur held in Bangladesh Jute Research Institute, Dhaka
Cooke RJ (1999) Modern methods for cultivar verification and the transgenic plant challenge. Seed Sci Technol 27:669–680
Nielsen G (1985) The use of isozymes as probes to identify and label plant varieties and cultivars, vol 12. Alan R. Liss, New York, pp 1–32
Joshi AB, Dhawan NL (1986) Genetic improvement of yield with special reference to self-fertilizing crops. Indian J Genet 26(1):101–113
Nevo E, Golenberg E, Beilies A, Brown AHD, Zohary D (1982) Genetic diversity and environmental association of wild wheat, Triticum diococcoides in Israel. Theor Appl Genet 62:241–254
Deng LQ, Li JQ, Li AQ (1994) Studies on the agronomic characters of kenaf germplasm and their utilization. China’s Fibre Crops 4:1–4 (in Chinese)
Degani C, Rowland LJ, Levi A, Hortynski JA, Galletta GJ (1998) DNA fingerprinting of strawberry (Fragaria X ananassa) cultivars using randomly amplified polymorphic DNA (RAPD) markers. Euphytica 102:247–253
Bowditch BM, Albright DG, Williams JGK, Braun MJ (1993) Use of randomly polymorphic DNA in comparative genome studies. Methods Genet Eng 224:294–309
Gupta PK, Balyan HS, Sharma PC, Ramesh B (1996) Microsatellites in plants: a new class of molecular markers. Curr Sci 70:45–54
Priolli RHG, Mendes-Junior CT, Arantes NE, Contel EPB (2002) Characterization of Brazilian soybean cultivars using microsatellite markers. Genet Mol Biol 25:185–193
Hokanson SC, Szewc-McFadden AK, Lamboy WF, McFerson JR (1998) Microsatellite (SSR) markers reveal genetic identities, genetic diversity and relationships in Malus × domestica borkh. Core subset collection. Theor Appl Genet 97:671–683
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Rajora OP, Rahman MH (2003) Microsatellite DNA and RAPD fingerprinting, identification and genetic relationships of hybrid poplar (Populus 3 canadensis) cultivars. Theor Appl Genet 106:470–477
Akter J, Islam MS, Sajib AA, Ashraf N, Haque S, Khan H (2008) Microsatellite markers for determining genetic identities and genetic diversity among jute cultivars. Aust J Crop Sci 1(3):97–107
Mir RR, Rustgi S, Sharma S, Singh R, Goyal A, Kumar J, Gaur A, Tyagi A, Khan H, Sinha MK, Balyan HS, Gupta PK (2008) A preliminary genetic analysis of fibre traits and the use of new genomic SSRs for genetic diversity in jute. Euphytica 161:413–427
Hossain MB, Haque S, Khan H (2002) DNA fingerprinting of jute germplasm by RAPD. J Biochem Mol Biol 35:414–419
Qi J, Zhou D, Wu W, Lin L, Fang P, Wu J (2003) The application of RAPD technology in genetic diversity detection of Jute. Ying Yon Sheng Tai XueBao 30:926–932
Haque S, Begum S, Sarker RH, Khan H (2007) Determining genetic diversity of some jute varieties and accessions using RAPD markers. Plant Tissue Cult Biotechnol 17(2):183–191
Roy A, Bandyopadhyay A, Mahapatra AK, Ghosh SK, Singh NK, Bansal KC, Koundal KR, Mohapatra T (2006) Evaluation of genetic diversity in jute (Corchorus species) using STMS, ISSR and RAPD markers. Plant Breed 125:292–297
Qi J, Zhou D, Wu W, Lin L, Wu J, Fang P (2003) Application of ISSR technology in genetic diversity detection of jute. Ying Yong Sheng Tai XueBao 14:1473–1477
Hossain MH, Awal A, Rahman MA, Haque S, Khan H (2003) Distinction between cold sensitive and -tolerant jute by DNA polymorphisms. J Biochem Mol Biol 35(5):427–432
Huq S, Islam, Sajib AA, Ashraf N, Haque S, Khan H (2009) Genetic diversity and relationship: in jute (Corchorus sp.) revealed by SSR markers. Bangladesh J Bot 38(2):153–161
Mir RR, Banerjee S, Das M, Gupta V, Tyagi AK, Sinha MK, Balyan HS, Gupta PK (2009) Development and characterization of large scale simple sequence repeats in jute. Crop Sci 49:1687–1694
Kundu Avijit, Sarkar Debabrata, Bhattacharjee Amit, Topdar Niladri, Sinha Mohit Kumar, Mahapatra Bikash Sinha (2011) A simple ethanol wash of the tissue homogenates recovers high-quality genomic DNA from Corchorus species characterized by highly acidic and proteinaceous mucilages. Electron J Biotechnol 14(15):1–8. doi:10.2225/vol14-issue1-fulltext-4
Sanguinetti CJ, Dias NE, Simpson AJG (1994) Rapid silver staining and recovery of PCR products separated on polyacrylamide gel. Biotechniques 17:915–919
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrell ME (1993) Optimizing parental selection for genetic linkage map. Genome 36:181–186
Das M, Banerjee S, Dhariwal R, Vyas S, Mir RR, Topdar N, Kundu A, Khurana JP, Tyagi AK, Sarkar D, Sinha MK, Balyan HS, Gupta PK (2012) Development of SSR markers and construction of a linkage map in jute. J Genet 91:1–11
Cooper HD, Spillan EC, Hodgkin T (2001) Broadening the genetic base of crops: an overview. Broadening the genetic base of crop production. CAB International, Wallingford, pp 1–23
Gupta PK, Rustgi S, Sharma S, Singh R, Kumar N, Balyan HS (2003) Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol Gen Genomics 270:315–323
Holton TA, Christopher JT, McClure L, Harker N, Henry RJ (2002) Identification and mapping of polymorphic SSR markers from expressed gene sequences of barley and wheat. Mol Breed 9:63–71
Ghosh RK, Wongkaew A, Sreewongchai T, Nakasathien S, Phumichai C (2014) Assessment of genetic diversity and population Structure in jute (Corchorus spp.) using simple sequence repeat (SSR) and amplified fragment length polymorphism (AFLP) markers. Nat Sci 48:83–94
Basu A, Ghosh M, Meyer R, Powell W, Basak SL, Sen SK (2004) Analysis of genetic diversity in cultivated jute determined by means of SSR markers and AFLP profiling. Crop Sci Soc Am 44:678–685
Acknowledgments
The authors highly acknowledge Indian Council of Agriculture Research (ICAR), India for providing financial support and facility to carry out the research work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ghosh, S., Meena, K., Sinha, M.K. et al. Genetic Diversity in Corchorus olitorius Genotypes Using Jute SSRs. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 87, 917–926 (2017). https://doi.org/10.1007/s40011-015-0652-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-015-0652-4