Abstract
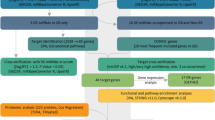
MicroRNAs (miRNAs) serve as critical modulators of post transcriptional gene regulation in response to stimuli such as narcotics. Hence, the analysis of miRNA expression may elucidate the molecular mechanisms underlying cellular response to morphine. Here, the expression profile of 750 known miRNAs was generated in BE (2)-C neuroblastoma cell line by using the miRCURY LNA microRNA qPCR array system, in order to identify morphine responsive miRNAs. This population of miRNAs can be used potentially to provide an insight into the functional mechanisms of this drug. Bioinformatics analysis was used to predict the target genes of morphine responsive miRNAs, and the altered expression of selected targets was investigated through quantitative polymerase chain reaction. The result showed that a total of 10 miRNAs were up-regulated, while 33 miRNAs were down-regulated in response to morphine treatment, considering at least 1.75 fold change in expression between treated and untreated cells. Among them, hsa-miR-29a-5p, has-miR-646, has-miR-1539, has-miR-412 and has-miR-937 showed the largest changes in expression level. Furthermore, the effect of multiple morphine responsive miRNAs on one target gene (cooperativity) was considered as the criterion to select target genes for the validation analysis. Cnr1, Jazf1, Nr2c2, Lonrf2, Bnc2, C1orf21 and eight more genes were selected based on cooperativity, and validated by using real time polymerase chain reaction.


Similar content being viewed by others
References
Spanagel R, Heilig M (2005) Addiction and its brain science. Addiction 100(12):1813–1822
Agrawal A, Lynskey MT (2008) Are there genetic influences on addiction: evidence from family, adoption and twin studies. Addiction 103(7):1069–1081
Ammon-Treiber S, Höllt V (2005) Morphine-induced Changes of Gene Expression in the Brain. Addict Biol 10(1):81–89
Shi J, Hui L, Xu Y, Wang F, Huang W, Hu G (2002) Sequence variations in the mu-opioid receptor gene (OPRM1) associated with human addiction to heroin. Hum Mutat 19(4):459–460
Standifer KM, Cheng J, Brooks AI, Honrado CP, Su W, Visconti LM, Biedler JL, Pasternak GW (1994) Biochemical and pharmacological characterization of mu, delta and kappa 3 opioid receptors expressed in BE (2)-C neuroblastoma cells. J Pharmacol Exp Ther 270(3):1246–1255
Bergeson SE, Helms ML, O’Toole LA, Jarvis MW, Hain HS, Mogil JS, Belknap JK (2001) Quantitative trait loci influencing morphine antinociception in four mapping populations. Mamm Genome 12(7):546–553
Kreek MJ, Zhou Y, Butelman ER, Levran O (2009) Opiate and cocaine addiction: from bench to clinic and back to the bench. Curr Opin Pharmacol 9(1):74–80
Wang J, Yuan W, Li MD (2011) Genes and pathways co-associated with the exposure to multiple drugs of abuse, including alcohol, amphetamine/methamphetamine, cocaine, marijuana, morphine, and/or nicotine: a review of proteomics analyses. Mol Neurobiol 44(3):269–286
Piechota M, Korostynski M, Sikora M, Golda S, Dzbek J, Przewlocki R (2012) Common transcriptional effects in the mouse striatum following chronic treatment with heroin and methamphetamine. Genes, Brain Behav 11(4):404–414
Doench JG, Sharp PA (2004) Specificity of microRNA target selection in translational repression. Genes Dev 18(5):504–511
Babashah S, Sadeghizadeh M, Tavirani MR, Farivar S, Soleimani M (2012) Aberrant microRNA expression and its implications in the pathogenesis of leukemias. Cell Oncol 35(5):317–334
Hausser J, Landthaler M, Jaskiewicz L, Gaidatzis D, Zavolan M (2009) Relative contribution of sequence and structure features to the mRNA binding of Argonaute/EIF2C–miRNA complexes and the degradation of miRNA targets. Genome Res. doi:10.1101/gr.091181.109
Miska EA (2005) How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev 15(5):563–568
Fiore R, Siegel G, Schratt G (2008) MicroRNA function in neuronal development, plasticity and disease. Biochim et Biophys Acta (BBA)-Gene Regul Mech 1779(8):471–478
Saba R, Schratt GM (2010) MicroRNAs in neuronal development, function and dysfunction. Brain Res 1338:3–13
Thomas M, Kalivas P, Shaham Y (2008) Neuroplasticity in the mesolimbic dopamine system and cocaine addiction. Br J Pharmacol 154(2):327–342
Chandrasekar V, Dreyer J-L (2009) microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol Cell Neurosci 42(4):350–362
Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65(1):55–63
Soong R, Ruschoff J, Tabiti K (2000) Detection of colorectal micrometastasis by quantitative RT-PCR of cytokeratin 20 mRNA. Roche Diagnostics internal publication, Lewes
Čikoš Š, Bukovská A, Koppel J (2007) Relative quantification of mRNA: comparison of methods currently used for real-time PCR data analysis. BMC Mol Biol 8(1):113
Karlen Y, McNair A, Perseguers S, Mazza C, Mermod N (2007) Statistical significance of quantitative PCR. BMC Bioinform 8(1):131
Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44(5):839–847
Wood EJ, Lipovich L (2012) MicroRNAs in opioid addiction: elucidating evolution. Front Genet. doi:10.3389/fgene.2012.00241
Renthal W, Nestler EJ (2009) Chromatin regulation in drug addiction and depression. Dialogues Clin Neurosci 11(3):257
Nguyen T, Kuo C, Nicholl MB, Sim M-S, Turner RR, Morton DL, Hoon D (2011) Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics 6(3):388–394
Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG (2009) DIANA-mirPath: integrating human and mouse microRNAs in pathways. Bioinformatics 25(15):1991–1993
Li C-Y, Mao X, Wei L (2008) Genes and (common) pathways underlying drug addiction. PLoS Comput Biol 4(1):e2
Mitra R, Bandyopadhyay S (2011) MultiMiTar: a novel multi objective optimization based miRNA-target prediction method. PLoS ONE 6(9):e24583
Brown JR, Sanseau P (2005) A computational view of microRNAs and their targets. Drug Discov Today 10(8):595–601
Zhou Y, Ferguson J, Chang JT, Kluger Y (2007) Inter-and intra-combinatorial regulation by transcription factors and microRNAs. BMC Genom 8(1):396
Kuhn DE, Martin MM, Feldman DS, Terry AV Jr, Nuovo GJ, Elton TS (2008) Experimental validation of miRNA targets. Methods 44(1):47–54
Nakajima T, Fujino S, Nakanishi G, Kim Y-S, Jetten AM (2004) TIP27: a novel repressor of the nuclear orphan receptor TAK1/TR4. Nucleic Acids Res 32(14):4194–4204
Acknowledgments
This work was financially supported by Tarbiat Modares University. The morphine used in the study was kindly provided by Health Ministry of Iran. The authors are deeply grateful to Babak Bakhshinejad for kindly reviewing the manuscript. The authors declare that there is no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sistani, R.N., Soltani, B.M. & Sadeghizadeh, M. Identifying microRNAs relating to morphine response in BE(2)-C cell line by microRNA profiling. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 87, 299–305 (2017). https://doi.org/10.1007/s40011-015-0614-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-015-0614-x