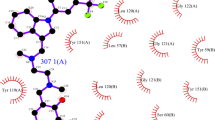
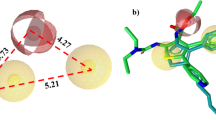
Abstract
TNF-α is a crucial cytokine in the process of inflammatory diseases. The adverse effect of TNF-α is mostly mediated by interaction of TNF-α with TNF-α receptor type I (TNFR1); therefore, discovery of molecules which can bind to TNFR1 preventing TNF-α-receptor complex formation would be of great interest. In the current study, using GRID/GOLPE program, a 3D-QSAR study was conducted on a series of synthetic TNFR1 binders, which resulted in a 3D-QSAR model with appropriate power of predictivity in internal (r2 = 0.94 and q2LOO = 0.74) and external (r2 = 0.66 and SDEP = 0.42) validations. The structural features of TNFR1 inhibitors essential for exerting activity were explored by analyzing the contour maps of the 3D-QSAR model showing that steric interactions and hydrogen bonds are responsible for exerting TNFR1 inhibitory activity. To propose potential chemical entities for TNFR1 inhibition, PubChem database was searched and the selected compounds were virtually tested for anti-TNFR1 activity using the generated model, resulting in two potential anti-TNFR1 compounds. Finally, the possible interactions of the compounds with TNFR1 were investigated using docking studies. The findings in the current work can pave the way for designing more potent anti-TNFR1 inhibitors.







Similar content being viewed by others
References
E.A. Carswell, L.J. Old, R.L. Kassel, S. Green, N. Fiore, B. Williamson, An endotoxin induced serum factor that cuases necrosis of tumors. Proc. Natl. Acad. Sci. USA. 72(9), 3666–3670 (1975)
I.A. Clark. How TNF was recognized as a key mechanism of disease. Cytokine Growth Factor Rev. 18(3–4), 335–343 (2007)
A. Ceramil, B. Beutler, The role of cachectin/TNF in endotoxic shock and cachexia. Immunol. Today. 9(1), 28–31 (1988)
P. Vandenabeele, W. Declercq, R. Beyaert, W. Fiers, Two tumour necrosis factor receptors: structure and function. Trends Cell. Biol. 5(10), 392–399 (1995)
K.J. Tracey, A. Cerami, Metabolic responses to cachectin/TNF. A brief review. Ann. N. Y. Acad. Sci. 587, 325–331 (1990)
M.J. Elliott, R.N. Maini, M. Feldmann, J.R. Kalden, C. Antoni, J.S. Smolen et al., Randomised double-blind comparison of chimeric monoclonal antibody to tumour necrosis factor α (cA2) versus placebo in rheumatoid arthritis. Lancet. 344(8930), 1105–1110 (1994)
M.A. Palladino, F.R. Bahjat, E.A. Theodorakis, L.L. Moldawer, Anti-TNF-α therapies: the next generation. Nat. Rev. Drug Discov. 2(9), 736–746 (2003)
W.J. Sandborn, S.B. Hanauer, S. Katz, M. Safdi, D.G. Wolf, R.D. Baerg et al., Etanercept for active Crohn’s disease: a randomized, double-blind, placebo-controlled trial. Gastroenterology. 121(5), 1088–1094 (2001)
D. Tracey, L. Klareskog, E.H. Sasso, J.G. Salfeld, P.P. Tak, Tumor necrosis factor antagonist mechanisms of action: a comprehensive review. Pharmacol. Therapeutics. 117(2), 244–279 (2008)
J.J. Gómez-Reino, L. Carmona, V. Rodríguez Valverde, E.M. Mola, M.D. Montero, Treatment of rheumatoid arthritis with tumor necrosis factor inhibitors may predispose to significant increase in tuberculosis risk: a multicenter active-surveillance report. Arthritis Rheum. 48(8), 2122–2127 (2003)
J.S. Lubel, A.G. Testro, P.W. Angus, Hepatitis B virus reactivation following immunosuppressive therapy: Guidelines for prevention and management. Int. Med. J. 37(10), 705–712 (2007)
C. Antoni, J. Braun, Side effects of anti-TNF therapy: current knowledge. Clin. Exp. Rheumatol. 20(6 SUPPL. 28), S-152-S-7 (2002)
S.A. Devos, N. Van Den Bossche, M. De Vos, J.M. Naeyaert, Adverse skin reactions to anti-TNF-alpha monoclonal antibody therapy. Dermatology. 206(4), 388–390 (2003)
R. Wolf, H. Matz, E. Orion, V. Ruocco, Anti-TNF therapies—the hope of tomorrow. Clin. Dermatol. 20(5), 522–530 (2002)
D. Faustman, M. Davis, TNF receptor 2 pathway: drug target for autoimmune diseases. Nat. Rev. Drug Discov. 9(6), 482–493 (2010)
R.M. Locksley, N. Killeen, M.J. Lenardo, The TNF and TNF receptor superfamilies: Integrating mammalian biology. Cell. 104(4), 487–501 (2001)
M. Leist, F. Gantner, S. Jilg, A. Wendel, Activation of the 55 kDa TNF receptor is necessary and sufficient for TNF- induced liver failure, hepatocyte apoptosis, and nitrite release. J. Immunol. 154(3), 1307–1316 (1995)
L. Mori, S. Iselin, G. De Libero, W. Lesslauer, Attenuation of collagen-induced arthritis in 55-kDa TNF receptor type 1 (TNFR1)-IgG1-treated and TNFR1-deficient mice. J. Immunol. 157(7), 3178–3182 (1996)
M.L. Olleros, R. Guler, N. Corazza, D. Vesin, H.P. Eugster, G. Marchal et al., Transmembrane TNF induces an efficient cell-mediated immunity and resistance to Mycobacterium bovis bacillus Calmette-Guérin infection in the absence of secreted TNF and lymphotoxin-α. J. Immunol. 168(7), 3394–3401 (2002)
B.M. Saunders, S. Tran, S. Ruuls, J.D. Sedgwick, H. Britton, B.J. Warwick, Transmembrane TNF is sufficient to initiate cell migration and granuloma formation and provide acute, but not long-term, control of Mycobacterium tuberculosis infection. J. Immunol. 174(8), 4852–4859 (2005)
Y. Mukai, T. Nakamura, M. Yoshikawa, Y. Yoshioka, S.I. Tsunoda, S. Nakagawa et al., Solution of the structure of the TNF-TNFR2 complex. Sci. Signal. 3(148), ra83 (2010)
T. Banno, A. Gazel, M. Blumenberg, Effects of tumor necrosis factor-α (TNFα) in epidermal keratinocytes revealed using global transcriptional profiling. J. Biol. Chem. 279(31), 32633–32642 (2004)
M.F. Rodrigues, C. Alves, B. Figueiredo, A.B. Rezende, S. Wohlres-Viana, Silva VLd et al., Tumour necrosis factor receptors and apoptosis of alveolar macrophages during early infection with attenuated and virulent Mycobacterium bovis. Immunology. 139(4), 503–512 (2013)
K.A. Zettlitz, V. Lorenz, K. Landauer, S. Münkel, A. Herrmann, P. Scheurich et al., ATROSAB, a humanized antagonistic anti-tumor necrosis factor receptor one-specific antibody. mAbs. 2(6), 639–647 (2010)
F.E. McCann, D.P. Perocheau, G. Ruspi, K. Blazek, M.L. Davies, M. Feldmann et al., Selective tumor necrosis factor receptor i blockade is antiinflammatory and reveals immunoregulatory role of tumor necrosis factor receptor II in collagen-induced arthritis. Arthritis Rheumatol. 66(10), 2728–2738 (2014)
P.H. Carter, P.A. Scherle, J.A. Muckelbauer, M.E. Voss, R.Q. Liu, L.A. Thompson et al., Photochemically enhanced binding of small molecules to the tumor necrosis factor receptor-1 inhibits the binding of TNF-α. Proc. Natl. Acad. Sci. USA. 98(21), 11879–11884 (2001)
M.E. Voss, P.H. Carter, A.J. Tebben, P.A. Scherle, G.D. Brown, L.A. Thompson et al., Both 5-arylidene-2-thioxodihydropyrimidine-4,6(1H,5H)-diones and 3-thioxo-2,3-dihydro-1H-imidazo[1,5-a]indol-1-ones are light-dependent tumor necrosis factor-α antagonists. Bioorg. Med. Chem. Lett. 13(3), 533–538 (2003)
J. Tong, Y. Wu, M. Bai, P. Zhan, 3D-QSAR and molecular docking studies on HIV protease inhibitors. J. Mol. Struct. 1129, 17–22 (2017)
A. Ghaleb, A. Aouidate, M. Ghamali, A. Sbai, M. Bouachrine, T. Lakhlifi, 3D-QSAR modeling and molecular docking studies on a series of 2,5 disubstituted 1,3,4-oxadiazoles. J. Mol. Struct. 1145, 278–284 (2017)
R.D. Cramer Iii, D.E. Patterson, J.D. Bunce, Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 110(18), 5959–5967 (1988)
P.J. Goodford, A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 28(7), 849–857 (1985)
R.C. Wade, K.J. Clark, P.J. Goodford, Further development of hydrogen bond functions for use in determining energetically favorable binding sites on molecules of known structure. 1. Ligand probe groups with the ability to form two hydrogen bonds. J. Med. Chem. 36(1), 140–147 (1993)
R.C. Wade, P.J. Goodford, Further development of hydrogen bond functions for use in determining energetically favorable binding sites on molecules of known structure. 2. Ligand probe groups with the ability to form more than two hydrogen bonds. J. Med. Chem. 36(1), 148–156 (1993)
M. Baroni, G. Costantino, G. Cruciani, D. Riganelli, R. Valigi, S. Clementi, Generating Optimal Linear PLS Estimations (GOLPE): An Advanced Chemometric Tool for Handling 3D-QSAR Problems. Quant. Struct. Activ. Relationsh. 12(1), 9–20 (1993)
HyperChem(TM) Professional 8.0.8; Hypercube, Inc, 1115 NW 4th Street, Gainesville, Florida 32601, USA, (2010)
C.B. Xue, X.T. Chen, X. He, J. Roderick, R.L. Corbett, B. Ghavimi et al., Synthesis and structure-activity relationship of a novel sulfone series of TNF-α converting enzyme inhibitors. Bioorg. Med. Chem. Lett. 14(17), 4453–4459 (2004)
N.L. Allinger. Conformational, analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms. J. Am. Chem. Soc. 99(25), 8127–8134 (1977)
Molecular Operating Environment (MOE), 2013.08; Chemical Computing Group Inc, 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7 (2016)
P. Goodford, GRID Molecular Discovery Ltd, Oxford, England (1995)
GOLPE 4.5, Multivariate Infometric Analysis; Srl., Viale dei Castagni 16 (Perugia, Italy, 1999)
M. Pastor, G. Cruciani, S. Clementi, Smart region definition: a new way to improve the predictive ability and interpretability of three-dimensional quantitative structure-activity relationships. J. Med. Chem. 40(10), 1455–1464 (1997)
G. Cruciani, K.A. Watson, Comparative molecular field analysis using GRID force-field and GOLPE variable selection methods in a study of inhibitors of glycogen phosphorylase b. J. Med. Chem. 37(16), 2589–2601 (1994)
K. Roy, S. Kar, P. Ambure, On a simple approach for determining applicability domain of QSAR models. Chemometr. Intell. Lab. Syst. 145, 22–29 (2015)
A.A. Alizadeh, M. Hamzeh-Mivehroud, B. Sokouti, S. Dastmalchi, An alignment-independent 3D-QSAR study on series of hydroxamic acid-based tumor necrosis factor-α converting enzyme inhibitors. J. Chemometr. 30(9), 537–547 (2016)
G. Jones, P. Willett, R.C. Glen, Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 245(1), 43–53 (1995)
G. Jones, P. Willett, R.C. Glen, A.R. Leach, R. Taylor, Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267(3), 727–748 (1997)
R.A. Laskowski, M.B. Swindells, LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inform. Model. 51(10), 2778–2786 (2011)
G.L. Wilson, M.A. Lill, Integrating structure-based and ligand-based approaches for computational drug design. Future Med. Chem. 3(6), 735–750 (2011)
C.H. Lee, H.C. Huang, H.F. Juan, Reviewing ligand-based rational drug design: the search for an ATP synthase inhibitor. Int. J. Mol. Sci. 12(8), 5304–5318 (2011)
O. Nicolotti, D. Gadaleta, G.F. Mangiatordi, M. Catto, A. Carotti, Applicability domain for QSAR models: where theory meets reality. Int. J. Quant. Struct. Prop. Relationsh. (IJQSPR). 1(1), 45–63 (2016) (2379–7487)
A. Golbraikh, A. Tropsha, Beware of q2! J. Mol. Graph. Model. 20(4), 269–276 (2002)
K. Roy, R.N. Das, P. Ambure, R.B. Aher, Be aware of error measures. Further studies on validation of predictive QSAR models. Chemometr. Intell. Lab. Syst. 152, 18–33 (2016)
C. Tintori, M. Magnani, S. Schenone, M. Botta, Docking, 3D-QSAR studies and in silico ADME prediction on c-Src tyrosine kinase inhibitors. Eur. J. Med. Chem. 44(3), 990–1000 (2009)
M. Gao, J. Skolnick, A comprehensive survey of small-molecule binding pockets in proteins. PLoS Comput. Biol. 9(10), e1003302 (2013)
Acknowledgements
The authors would like to thank the Research Office and Biotechnology Research Center of Tabriz University of Medical Sciences for providing financial and facility support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sharifi, M., Alizadeh, A.A., Hamzeh-Mivehroud, M. et al. Computational explorations to gain insight into the structural features of TNF-α receptor I inhibitors. J IRAN CHEM SOC 15, 2519–2531 (2018). https://doi.org/10.1007/s13738-018-1440-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-018-1440-x