Abstract
Breast cancer is an expanding threat that leads to many women's death worldwide. Despite the improvement of the early detection methods and treatment, still, there is a high number of breast cancer mortality. To increase patient survival in breast cancer, identifying novel biomarkers is essential for therapeutics targets. The Glycophorin C (GYPC) gene is correlated with patient survival, which can be a possible biomarker for early detection in breast cancer progression. However, the expression of GYPC is not clearly defined in breast cancer. Here, we widely analyzed the expression pattern of GYPC in breast cancer and patient survival datasets through several bioinformatics tools. GYPC mRNA expression using ONCOMINE, GENT2, and GTX2 webs. Also, The co-expression profile of GYPC has been repossessed from Ma breast four datasets from Oncomine dataset. Our study revealed that mRNA expression of GYPC is strongly correlated with the survival of breast cancer patients, suggesting its role as a tumor suppressor. The downregulation of GYPC in breast cancer tissue is examined by promoter methylation and copy number alterations. The downregulation of GYPC expression was significantly correlated with high patient survival. Moreover, we performed pathway analysis via Enricher and gene ontology web using 20 positively correlated genes. Consequently, our analyzed data suggested that GYPC might be an essential therapeutics and prognostic biomarker in breast cancer.









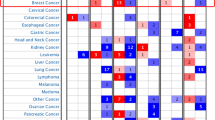
Source databases for KM plots include GEO, EGA, and TCGA



Similar content being viewed by others
Data availability
The authors declare that all the data supporting the findings of this study are available within the paper and supplementary file.
References
Barman UD, Saha SK, Kader MA, Jamal MAHM, Sharma SP, Samad A, Rahman MS (2020) Clinicopathological and prognostic significance of GPC3 in human breast cancer and its 3D structure prediction. Netw Model Anal Health Inf Bioinform 9:1–18
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV, Varambally S (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19:649–658
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform 14:128
Chen L, Chu C, Lu J, Kong X, Huang T, Cai Y-D (2015) Gene ontology and KEGG pathway enrichment analysis of a drug target-based classification system. PLoS ONE 10:e0126492
Cornen S, Guille A, Adélaïde J, Addou-Klouche L, Finetti P, Saade M-R, Manai M, Carbuccia N, Bekhouche I, Letessier A, Raynaud S, Charafe-Jauffret E, Jacquemier J, Spicuglia S, de The H, Viens P, Bertucci F, Birnbaum D, Chaffanet M (2014) Candidate luminal B breast cancer genes identified by genome, gene expression and DNA methylation profiling. PLoS ONE 9:e81843
Ehrlich M (2019) DNA hypermethylation in disease: mechanisms and clinical relevance. Epigenetics 14:1141–1163
Farkas SA, Milutin-Gašperov N, Grce M, Nilsson TK (2013) Genome-wide DNA methylation assay reveals novel candidate biomarker genes in cervical cancer. Epigenetics 8:1213–1225
Feng L, Jin F (2018) Screening of differentially methylated genes in breast cancer and risk model construction based on TCGA database. Oncol Lett 16:6407–6416
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:11
Geng H, Neuberg D, Paietta E, Deng X, Li Y, Xin Y, Racevskis J, Ketterling R, Richards SM, Tallman MS, Rowe JM, Litzow MR, Elemento O, Melnick AM (2010) Integrative genome-wide DNA methylation and gene expression analysis reveals biological and clinical insights in adult acute lymphoblastic leukemia. Blood 116:852–852
Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, Zhao L, Zhang Y, Huang B, Lu J (2014) LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis 5:e1287–e1287
Jézéquel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, Campion L (2012) bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat 131:765–775
Jézéquel P, Frénel J-S, Campion L, Guérin-Charbonnel C, Gouraud W, Ricolleau G, Campone M (2013) bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database. https://doi.org/10.1093/database/bas060
Kazanietz MG, Caloca MJ (2017) The Rac GTPase in cancer: from old concepts to new paradigms. Can Res 77:5445
Kim J, Piao H-L, Kim B-J, Yao F, Han Z, Wang Y, Xiao Z, Siverly AN, Lawhon SE, Ton BN (2018) Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet 50:1705–1715
Kleczko EK, Kwak JW, Schenk EL, Nemenoff RA (2019) Targeting the complement pathway as a therapeutic strategy in lung cancer. Front Immunol. https://doi.org/10.3389/fimmu.2019.00954
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44:W90-97
Le Van Kim C, Piller V, Cartron J, Colin Y (1996) Glycophorins C and D are generated by the use of alternative translation initiation sites. Blood. https://doi.org/10.1182/blood.V88.6.2364.bloodjournal8862364
Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y, Weinstein JN, Liang H (2015) TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Can Res 75:3728–3737
Li Z, Hou P, Fan D, Dong M, Ma M, Li H, Yao R, Li Y, Wang G, Geng P (2017) The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ 24:59–71
Lisany NF, Jamal MAHM, Chung H-J, Hong S-T, Rahman MS (2020) Prognostic significance of the Cdk5 gene in breast cancer: an in silico study. Netw Model Anal Health Inform Bioinform 9:1–10
Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D (2015) A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27:370–381
Liu Y, Hu Z, Yang C, Wang S, Wang W, Zhang Q (2017) A post-genome-wide association study validating the association of the glycophorin C gene with serum hemoglobin level in pig. Asian Australas J Anim Sci 30:638–642
Liu L, Zhang Y, Lu J (2020) The roles of long noncoding RNAs in breast cancer metastasis. Cell Death Dis 11:749
Malvia S, Bagadi SAR, Pradhan D, Chintamani C, Bhatnagar A, Arora D, Sarin R, Saxena S (2019) Study of gene expression profiles of breast cancers in Indian women. Sci Rep 9:10018
Martin JC, Herbert BS, Hocevar BA (2010) Disabled-2 downregulation promotes epithelial-to-mesenchymal transition. Br J Cancer 103:1716–1723
Mi H, Thomas P (2009) PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. In: Nikolsky Y, Bryant J (eds) Protein networks and pathway analysis. Methods in Molecular Biology (Methods and Protocols), vol 563. Humana Press. https://doi.org/10.1007/978-1-60761-175-2_7
Nagy Á, Lánczky A, Menyhárt O, Győrffy B (2018) Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 8:1–9
Ning C, Li YY, Wang Y, Han GC, Wang RX, Xiao H, Li XY, Hou CM, Ma YF, Sheng DS, Shen BF, Feng JN, Guo RF, Li Y, Chen GJ (2015) Complement activation promotes colitis-associated carcinogenesis through activating intestinal IL-1beta/IL-17A axis. Mucosal Immunol 8:1275–1284
Ojo D, Rodriguez D, Wei F, Bane A, Tang D (2019) Downregulation of CYB5D2 is associated with breast cancer progression. Sci Rep 9:6624
Paramita S, Raharjo EN, Niasari M, Azizah F, Hanifah NA (2019) Luminal B is the most common intrinsic molecular subtypes of invasive ductal breast carcinoma patients in East Kalimantan, Indonesia. Asian Pac J Cancer Prev 20:2247–2252
Park S-J, Yoon B-H, Kim S-K, Kim S-Y (2019) GENT2: an updated gene expression database for normal and tumor tissues. BMC Med Genomics 12:101
Pomaznoy M, Ha B, Peters B (2018) GOnet: a tool for interactive gene ontology analysis. BMC Bioinform 19:470
Rahman MF, Rahman MR, Islam T, Zaman T, Shuvo MAH, Hossain MT, Islam MR, Karim MR, Moni MA (2019) A bioinformatics approach to decode core genes and molecular pathways shared by breast cancer and endometrial cancer. Inform Med Unlocked 17:100274
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6:1
Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P (2007) Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9:166
Saha SK, Biswas PK, Gil M, Cho S-G (2019) High expression of TTYH3 is related to poor clinical outcomes in human gastric cancer. J Clin Med 8:1762
Saha SK, Kader MA, Samad KA, Biswas KC, Rahman MA, Parvez MAK, Rahman MS (2020) Prognostic and clinico-pathological significance of BIN1 in breast cancer. Inform Med Unlocked 19:100327
Sen MK, Almuslehi MS, Gyengesi E, Myers SJ, Shortland PJ, Mahns DA, Coorssen JR (2019) Suppression of the peripheral immune system limits the central immune response following cuprizone-feeding: relevance to modelling multiple sclerosis. Cells 8:1314
Smith JC, Sheltzer JM (2018) Systematic identification of mutations and copy number alterations associated with cancer patient prognosis. Elife 7:e39217
Suriyamurthy S, Baker D, ten Dijke P, Iyengar PV (2019) Epigenetic reprogramming of TGF-β signaling in breast cancer. Cancers 11:726
Tang Z, Kang B, Li C, Chen T, Zhang Z (2019) GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 47:W556–W560
Tendl KA, Bago-Horvath Z (2020) Molecular profiling in breast cancer—ready for clinical routine? MEMO. https://doi.org/10.1007/s12254-020-00578-0
Vadrevu SK, Chintala NK, Sharma SK, Sharma P, Cleveland C, Riediger L, Manne S, Fairlie DP, Gorczyca W, Almanza O, Karbowniczek M, Markiewski MM (2014) Complement c5a receptor facilitates cancer metastasis by altering T-cell responses in the metastatic niche. Can Res 74:3454–3465
Wang Y, Song B, Zhu L, Zhang X (2019) Long non-coding RNA, LINC01614 as a potential biomarker for prognostic prediction in breast cancer. PeerJ 7:e7976
Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, Park PK, Qin L, Wei Y, Hawke DH (2014) lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 159:1110–1125
Xu S, Zhu J, Wu Z (2014) Loss of Dab2 expression in breast cancer cells impairs their ability to deplete TGF-β and induce tregs development via TGF-β. PLoS ONE 9:e91709
Xu H, Moni MA, Liò P (2015) Network regularised Cox regression and multiplex network models to predict disease comorbidities and survival of cancer. Comput Biol Chem 59:15–31
Yu Y, Lv F, Liang D, Yang Q, Zhang B, Lin H, Wang X, Qian G, Xu J, You W (2017) HOTAIR may regulate proliferation, apoptosis, migration and invasion of MCF-7 cells through regulating the P53/Akt/JNK signaling pathway. Biomed Pharmacother 90:555–561
Zhang C, Zhao H, Li J, Liu H, Wang F, Wei Y, Su J, Zhang D, Liu T, Zhang Y (2015) The identification of specific methylation patterns across different cancers. PLoS ONE 10:e0120361
Zhang Y, Fang L, Zang Y, Xu Z (2018) Identification of core genes and key pathways via integrated analysis of gene expression and DNA methylation profiles in bladder cancer. Med Sci Monit 24:3024–3033
Zhang R, Liu Q, Li T, Liao Q, Zhao Y (2019) Role of the complement system in the tumor microenvironment. Cancer Cell Int 19:300
Zhu GZ, Yang YL, Zhang YJ, Liu W, Li MP, Zeng WJ, Zhao XL, Chen XP (2017) High expression of AHSP, EPB42, GYPC and HEMGN predicts favorable prognosis in FLT3-ITD-negative acute myeloid leukemia. Cell Physiol Biochem 42:1973–1984
Zinia JA, Rahman MS (2020) Evaluation of the prognostic significance of CDK6 in breast cancer. Netw Modeling Anal Health Inform Bioinform 9:40
Funding
MSR acknowledges that this research was supported by grants from the Research & Development Project from the Ministry of Science and Technology and Jashore University of Science and Technology, Bangladesh.
Author information
Authors and Affiliations
Contributions
MSR conceived the idea, analyzed the data, and drafted the manuscript. PKB, SKS, and MAM reviewed the manuscript. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This in silico study does not involve human, animal, plant, or clinical experiments. Thus, it does not require ethical approval.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rahman, M., Biswas, P.K., Saha, S.K. et al. Identification of glycophorin C as a prognostic marker for human breast cancer using bioinformatic analysis. Netw Model Anal Health Inform Bioinforma 11, 7 (2022). https://doi.org/10.1007/s13721-021-00352-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-021-00352-0