Abstract
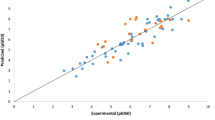
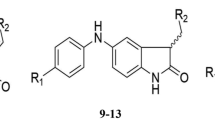
The pGI50 cytotoxicity values of 112 compounds on K-562 cancer cell line were modeled to illustrate the quantitative structure–activity relationship (QSAR) of the compounds. The dataset were divided into training and test set through Kennard-stone algorithm, while the pool of molecular descriptors calculated with paDEL descriptor metric program was subjected to the genetic functional algorithm (GFA) for selection of descriptor to be modeled. The best QSAR model developed was then subjected to a rigorous statistical test. The statistical significance of the model was verified by calculating the values of Q2LOO (0.845), Q2F1 (0.9397), Q2F2 (0.6862) and R2pred (0.6862) needed to evaluate the strength and robustness of the model. The result of the internal and external validation of the model indicates that the model is good and could be used to predict the GI50 of anticancer compounds on K-562 leukemia cell line. The model developed was used in designing new anticancer drugs with higher activity or more potent and less toxic in nature when compared to the lead compound. These compounds significant activities were found to correlate to with some of the molecular descriptors such as the number of hydrogen bond acceptors present in the surface of the molecule.



Similar content being viewed by others
References
Andersson LC, Nilsson K, Gahmberg CG (1979) K562—a human erythroleukemic cell line. Int J Cancer 23(2):143–147
Broto P, Devillers J (1990) Autocorrelation of properties distributed on molecular graphs. Kluwer Academic Publishers, Dordrecht
Broto P, Moreau G, Vandycke C (1984) Molecular structures: perception, autocorrelation descriptor and sar studies: system of atomic contributions for the calculation of the n-octanol/water partition coefficients. Eur J Med Chem 19(1):71–78
Chopade SM, Phadnis PP, Hodage AS, Wadawale A, Jain VK (2015) Synthesis, characterization, structures and cytotoxicity of platinum(II) complexes containing dimethylpyrazole based selenium ligands. Inorg Chim Acta 427:72–80. https://doi.org/10.1016/j.ica.2014.11.017
Denton P (2001) Generating coursework feedback for large groups of students using MS Excel and MS Word. Univ Chem Educ 5(1):1–8
Dimić D, Mercader AG, Castro EA (2015) Chalcone derivative cytotoxicity activity against MCF-7 human breast cancer cell QSAR study. Chemometr Intell Lab Syst 146:378–384. https://doi.org/10.1016/j.chemolab.2015.06.011
Drexler HG (2000) The leukemia-lymphoma cell line factsbook. Academic Press, Cambridge
Evans DA (2014) History of the Harvard ChemDraw project. Angew Chem Int Ed 53(42):11140–11145
Fan Y, Lu H, An L, Wang C, Zhou Z, Feng F, Zhao Q et al (2016) Effect of active fraction of Eriocaulon sieboldianum on human leukemia K562 cells via proliferation inhibition, cell cycle arrest and apoptosis induction. Environ Toxicol Pharmacol 43:13–20
Fatemi MH, Heidari A, Gharaghani S (2015) QSAR prediction of HIV-1 protease inhibitory activities using docking derived molecular descriptors. J Theor Biol 369:13–22. https://doi.org/10.1016/j.jtbi.2015.01.008
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127(12):2893–2917
Golbraikh A, Tropsha A (2002) Beware of q 2! J Mol Graph Model 20(4):269–276
Gramatica P, Giani E, Papa E (2007) Statistical external validation and consensus modeling: a QSPR case study for Koc prediction. J Mol Graph Model 25(6):755–766
Hall LH, Kier LB (1995) Electrotopological state indices for atom types: a novel combination of electronic, topological, and valence state information. J Chem Inf Comput Sci 35(6):1039–1045
Hehre WJ, Huang WW (1995) Chemistry with computation: an introduction to SPARTAN. Wavefunction, Inc, Irvine
Iuliano A, Strianese D, Uccello G, Diplomatico A, Tebaldi S, Bonavolontà G (2012) Risk factors for orbital exenteration in periocular basal cell carcinoma. Am J Ophthalmol 153(2):238–241.e231
Kar S, Roy K (2012) QSAR of phytochemicals for the design of better drugs. Expert Opin Drug Discov 7(10):877–902. https://doi.org/10.1517/17460441.2012.716420
Karimiani EG, Marriage F, Merritt AJ, Burthem J, Byers RJ, Day PJ (2014) Single-cell analysis of K562 cells: an imatinib-resistant subpopulation is adherent and has upregulated expression of BCR-ABL mRNA and protein. Exp Hematol 42(3):183–191e185
Kennard RW, Stone LA (1969) Computer aided design of experiments. Technometrics 11(1):137–148
Li Z, Wan H, Shi Y, Ouyang P (2004) Personal experience with four kinds of chemical structure drawing software: review on ChemDraw, ChemWindow, ISIS/Draw, and ChemSketch. J Chem Inf Comput Sci 44(5):1886–1890
Liu S, Cao C, Li Z (1998) Approach to estimation and prediction for normal boiling point (NBP) of alkanes based on a novel molecular distance-edge (MDE) vector λ. J Chem Inf Comput Sci 38(3):387–394
Lozzio CB, Lozzio BB (1975) Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood 45(3):321–334
Marx KA, O’Neil P, Hoffman P, Ujwal M (2003) Data mining the NCI cancer cell line compound GI50 values: identifying quinone subtypes effective against melanoma and leukemia cell classes. J Chem Inf Comput Sci 43(5):1652–1667
Moreau G, Broto P (1980a) The auto-correlation of a topological-structure—a new molecular descriptor, vol 4. Gauthier-Villars, Paris Cedex, pp 359–360
Moreau G, Broto P (1980b) Auto-correlation of molecular-structures, application to SAR studies. Nouveau J Chim N J Chem 4(12):757–764
News B (2003) Cancer number one killer of men. Health. http://news.bbc.co.uk/2/hi/health/3019801.stm. Accessed 18 Apr 2016
Ojha PK, Mitra I, Das RN, Roy K (2011) Further exploring r m 2 metrics for validation of QSPR models. Chemometr Intell Lab Syst 107(1):194–205
Parkin DM, Boyd L, Walker L (2011) 16. The fraction of cancer attributable to lifestyle and environmental factors in the UK in 2010. Br J Cancer 105:S77–S81
Rajer-Kanduč K, Zupan J, Majcen N (2003) Separation of data on the training and test set for modelling: a case study for modelling of five colour properties of a white pigment. Chemometr Intell Lab Syst 65(2):221–229
Roy K, Chakraborty P, Mitra I, Ojha PK, Kar S, Das RN (2013) Some case studies on application of “rm2” metrics for judging quality of quantitative structure–activity relationship predictions: emphasis on scaling of response data. J Comput Chem 34(12):1071–1082
Roy K, Kar S, Das RN (2015a) A primer on QSAR/QSPR modeling: fundamental concepts. Springer, Berlin
Roy K, Kar S, Das RN (2015b) Understanding the basics of QSAR for applications in pharmaceutical sciences and risk assessment. Academic Press, Cambridge
Roy K, Kar S, Ambure P (2015c) On a simple approach for determining applicability domain of QSAR models. Chemometr Intell Lab Syst 145:22–29. https://doi.org/10.1016/j.chemolab.2015.04.013
Sabet R, Mohammadpour M, Sadeghi A, Fassihi A (2010) QSAR study of isatin analogues as in vitro anti-cancer agents. Eur J Med Chem 45(3):1113–1118
Saptoro A, Tadé MO, Vuthaluru H (2012) A modified Kennard-Stone algorithm for optimal division of data for developing artificial neural network models. Chem Prod Process Model 7(1):1–14
Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA 65(1):5–29
Speck-Planche A, Kleandrova VV, Luan F, Cordeiro MND (2012a) Rational drug design for anti-cancer chemotherapy: multi-target QSAR models for the in silico discovery of anti-colorectal cancer agents. Bioorg Med Chem 20(15):4848–4855
Speck-Planche A, Kleandrova VV, Luan F, Cordeiro MNDS. (2012b) Chemoinformatics in anti-cancer chemotherapy: multi-target QSAR model for the in silico discovery of anti-breast cancer agents. Eur J Pharm Sci 47(1):273–279. https://doi.org/10.1016/j.ejps.2012.04.012
Supratik Kar KR (2010) Development and validation of a robust QSAR model for prediction of carcinogenicity of drugs. Indian J Biochem Biophys 48:111–122
Todeschini R, Consonni V (2009) Molecular descriptors for chemoinformatics, vol 41. Wiley, Hoboken
Tropsha A (2010) Best practices for QSAR model development, validation, and exploitation. Mol Inf 29(6–7):476–488
Tropsha A, Gramatica P, Gombar VK (2003) The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. QSAR Comb Sci 22(1):69–77
Viswanadhan VN, Ghose AK, Revankar GR, Robins RK (1989) Atomic physicochemical parameters for three dimensional structure directed quantitative structure–activity relationships. 4. Additional parameters for hydrophobic and dispersive interactions and their application for an automated superposition of certain naturally occurring nucleoside antibiotics. J Chem Inf Comput Sci 29(3):163–172
World Health Organization (2002) National cancer control programmes: policies and managerial guidelines. World Health Organization
Wu X, Fini P, Keller S, Tarsa E, Heying B, Mishra U, Speck J et al (1996) Morphological and structural transitions in GaN films grown on sapphire by metal-organic chemical vapor deposition. Jpn J Appl Phys 35(12B):L1648
Yap CW (2011) PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem 32(7):1466–1474
Acknowledgements
We would like to acknowledge the National Cancer institute for providing the material data used for the QSAR study in the website (https://wiki.nci.nih.gov/display/NCIDTPdata/NCI-60+Growth+Inhibition+Data).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Arthur, D.E., Uzairu, A., Mamza, P. et al. In silico modelling of quantitative structure–activity relationship of multi-target anticancer compounds on k-562 cell line. Netw Model Anal Health Inform Bioinforma 7, 11 (2018). https://doi.org/10.1007/s13721-018-0168-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-018-0168-y