Abstract
Key message
Increasing human impacts on forests, including unintentional movement of pathogens, climate change, and large-scale intensive plantations, are associated with an unprecedented rate of new diseases. An evolutionary ecology perspective can help address these challenges and provide direction for sustainable forest management.
Context
Forest pathology has historically relied on an ecological approach to understand and address the practical management of forest diseases. A widening of this perspective to include evolutionary considerations has been increasingly developed in response to the rising rates of genetic change in both pathogen populations and tree populations due to human activities.
Aims
Here, five topics for which the evolutionary perspective is especially relevant are highlighted.
Results
The first relates to the evolutionary diversity of fungi and fungal-like organisms, with issues linked to the identification of species and their ecological niches. The second theme deals with the evolutionary processes that allow forest pathogens to adapt to new hosts after introductions or to become more virulent in homogeneous plantations. The third theme presents issues linked to disease resistance in tree breeding programs (e.g., growth-defense trade-offs) and proposes new criteria and methods for more durable resistance. The last two themes are dedicated to the biotic environment of the tree–pathogen system, namely, hyperparasites and tree microbiota, as possible solutions for health management.
Conclusion
We conclude by highlighting three major conceptual advances brought by evolutionary biology, i.e., that (i) “not everything is everywhere”, (ii) evolution of pathogen populations can occur on short time scales, and (iii) the tree is a multitrophic community. We further translate these into a framework for immediate policy recommendations and future directions for research.
Similar content being viewed by others
1 Introduction: a brief history of forest pathology and its primary concepts
Forest pathology deals with the diseases of forest trees, which are mainly caused by fungal and oomycete pathogens, in both their fundamental and applied aspects. As such, the history of forest pathology has been shaped not only by the progress of ideas and concepts in the more general disciplines of microbiology and health sciences but also by changes in the practice of forestry. Forest pathology has obvious common roots with plant pathology, but is not simply one of its specialities. Long time scales, diversity, and heterogeneity are hallmarks of forests, in both their biological and socio-economic dimensions. Forest trees are still mostly nondomesticated species, highly diverse, growing in richly biotic and heterogeneous environments. Until recently, forest products have been harvested for the greatest part from natural or seminatural forests, with rotation periods generally exceeding 50 years (FAO 2010). As a consequence, forest pathology, as a speciality of forestry, has had a tradition of systemic and long-term thinking and, probably more so and earlier than plant pathology, an ecological approach. Integrating an evolutionary perspective is a natural extension, especially in the context of global changes.
Robert Hartig, the author of two forest pathology textbooks in 1874 and 1882, is generally recognized as the father of forest pathology, following in Anton de Bary’s footsteps, who is considered to be the father of plant pathology (Kutschera and Hossfeld 2012). In support to the germ theory of disease, de Bary provided experimental evidence of the role of Phytophthora infestans in potato late blight epidemics, refuting the old theory of spontaneous generation, while Hartig proposed a new concept of tree decay whereby fungi were the cause and not the consequence of the degradation (Merrill and Shigo 1979). The identification and biological characterization of the causal agents of the diseases affecting trees were still the primary objectives of forest pathologists at the end of the nineteenth and the beginning of the twentieth centuries. From the very beginning, forest pathologists were concerned with both productivity (fungi affecting wood production and preservation) and conservation issues (pathogens threatening natural resources) (Peterson and Griffith 1999). As the potato late blight epidemics did for plant pathology, severe outbreaks caused by the introduction of exotic pathogens have marked the history of forest pathology, such as chestnut blight and white pine blister rust in North America and Europe (Anagnostakis 1987; Delatour et al. 1985). Forest health has also been recurrently marked by regionally important declines affecting a variety of species, e.g., oaks in Europe or maple in North America (Delatour 1983; Tainter and Baker 1996). These phenomena, leading to progressive debilitation of trees and ultimately mortality, highlight the potential long-term effects of weakening factors, especially soil and climate, interacting with the genetic background of trees. As early as 1963, George (Hepting 1963) wrote that “field crops [mostly annual plants]…reflect weather changes; whereas trees […] will reflect, in addition, climate changes.” The “tree decline concept,” with biotic and abiotic factors acting and interacting in sequence as predisposing, inciting, or contributing factors was later formalized by Manion (1981). More generally, forest pathologists have long been interested in and have studied the relationships between environment, e.g., local site factors, and disease. In contrast to more intensively managed agricultural systems, forest environments are heterogeneous, and their natural constraints (fertility, water availability) are not usually overcome by external, human inputs. Moreover, direct control of diseases through fungicide applications has usually not been considered to be necessary or appropriate, for reasons such as cost, practical feasibility, and environmental concerns. Disease management in forests has thus traditionally been viewed as necessarily based on “good” silvicultural practices and preventive actions deriving from the knowledge of risk factors. Hazard-rating systems, which identify sites where pathogens are likely to have a high impact and which should thus be avoided for the planting of susceptible species, are examples of successful applications of forest epidemiology (Tainter and Baker 1996).
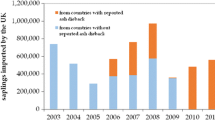
The issues faced by forest pathologists today would not be so different than those addressed by their predecessors, but for the dramatic acceleration and intensity of changes. The apparition of new diseases caused by alien invasive pathogens is rapidly increasing with the intensification of international trade and travels (Brasier 2008; Desprez-Loustau et al. 2010; Pyšek et al. 2010; Santini et al. 2013; Roy et al. 2014), presenting a major challenge to forest pathology today. Meanwhile, climate change is expected to affect the probability of establishment of introduced pathogens and more generally to affect the distribution and severity of diseases (e.g., Shaw and Osborne 2011; Sturrock et al. 2011). Indeed, forest pathologists pioneered the use of risk mapping for plant disease under climate change by combining epidemiological models and climate change scenarios (Brasier 1996; Booth et al. 2000; Bergot et al. 2004). The rapid intensification of forestry is likewise influencing forest pathology. The increasing demand for wood, biomass, fuel, and other forest-derived products has led to a large expansion of planted forests, by around 5 million hectares per year on average in the last decade, with the aim to increase productivity (FAO 2010). In some countries, especially in the southern hemisphere and Southeast Asia, where forest industries have implemented large-scale, management-intensive monocultures of Eucalyptus, Acacia, and Pinus, plantations currently account for most of the wood supply (Paquette and Messier 2009). However, new disease emergences threaten productivity in these new forest ecosystems, as has been reported for stem cankers and leaf and shoot blight of Eucalypts or root rots of Acacia spp. (Su-See 1999; Mohan and Manokaran 2013; Liebhold and Wingfield 2014). The strong impact of pathogen introductions and forest management practices on forest diseases can be illustrated by trends observed in the database of the French Forest Health service (Fig. 1).
The database of the French Health Service includes approximately 60,000 reports, from 1989 to 2014, which provides a general view of all pathogens (and other causes) affecting forest health. A total of 24,375 records have been identified at the pathogen species level, for a total of 231 species, among which 29 introduced species for 8634 records and 202 nonintroduced species for 15 741 records. a Introduced pathogens are overrepresented among the most frequent forest diseases; Fisher’s exact test ; **p = 0.0014. The value above each column indicates the number of pathogen species. The 19 more frequent pathogens represent 81% of all disease records. b The part of reports associated with introduced pathogens has much increased in the recent period especially since the detection of Hymenoscyphus fraxineus in 2008 in eastern France. c A much higher rate of reports is observed in forest plantations compared to less intensively managed forests (1989–2006 data). This probably includes both higher pathogen load but also higher attention paid to growth losses in these systems (lower tolerance to damage by the forester)
Long before disease regulation was formally recognized as an ecosystem service (Millennium Ecosystem Assessment 2005), forest pathology took an empirical approach to the study of the ecological processes that regulate forest health. The overall low level of disease in natural and seminatural forests, with a few exceptions of devastating native diseases (Hansen and Goheen 2000), is consistent with the general observation that native plant populations rarely suffer from devastating epidemics, contrary to what is observed in crops without fungicide applications (Jousimo et al. 2014). A major exception to the effectiveness of natural disease regulation, at least in the short term, is constituted by diseases caused by some exotic pathogens. The awareness of the increasing impacts of human activities on ecosystems, including forests, has emphasized the need for global approaches and has changed our views on evolution of species. It has been increasingly realized that evolutionary changes can be observed at short time-scales, especially in ecosystems under strong anthropogenic influence (Palumbi 2001; Burdon and Thrall 2008; Jousimo et al. 2014). Thus, the traditional view considering evolutionary biology and ecology as two independent fields has changed dramatically (Koch et al. 2014). The speed of adaptation of organisms to keep pace with the human alterations of the environment has become a matter of concern (Carroll et al. 2014). In particular, the ever-faster pace of disease emergences and the severity of the damage they cause has emphasized the need to better understand and take into consideration the coevolutionary processes between pathogens and their host tree populations (Parker and Gilbert 2004; Schoettle and Sniezko 2007; Hendry et al. 2011; Ennos 2015). At the same time, the revolution brought by DNA technologies and their reducing costs has opened new prospects for population genetic and environmental metagenomic studies (Neale and Ingvarsson 2008; Di Bella et al. 2013), which can shed new light on the ecology and evolution of forest diseases and open new avenues for disease control.
In the current context of human-induced accelerated changes, applying evolutionary principles to disease and pest management has been advocated by several authors as the way forward to slow the pathogen arms race in human, animal, or plant diseases, (Combes 2001; Palumbi 2001; Varki 2012; Vander Wal et al. 2014). Anton de Bary himself referred to Darwin’s principle of natural selection when defining plant–parasite interactions (Kutschera and Hossfeld 2012). Indeed, evolutionary biology provides the concepts and methods to understand the diversity and adaptive characteristics of organisms, thus allowing the identification of pathogens through phylogenetics, and the prediction and even manipulation of selection and adaptation processes involved in immune or pathogenic functions (Carroll et al. 2014). The idea that “nothing in forest pathology makes sense except in the light of evolution,” paraphrasing Dobzhansky, could now seem obvious and not deserving of further development (Varki 2012). Many recent articles dealing with forest pathology indeed have an evolutionary background (e.g., Hansen and Goheen 2000; Pinon and Frey 2005; Kinloch et al. 2008; Garbelotto et al. 2010; Hayden et al. 2011; Cruikshank and Jaquish 2014; Franceschini et al. 2014). However, we contend that an evolutionary perspective is still not sufficiently emphasized and applied to the management of forest diseases. For example, evolutionary considerations, especially relating to interactions with pathogens, are still ignored in a recently proposed “novel tree breeding strategy” advocating domestication of forest trees in a changing world (Harfouche et al 2012). In line with other recent initiatives in forest pathology (e.g., Cavers 2015; Cavers and Cottrell 2015) and taking benefit of insights from other fields (plant pathology, invasion ecology, ecology of parasitism, etc.), this article aims to show how an evolutionary ecology perspective can improve and even change our understanding of forest diseases and help address the challenges of today and tomorrow. Rather than a comprehensive review of recent developments in forest pathology, we highlight and focus on some key topics, for which the evolutionary perspective is especially relevant and could provide new directions for forest research or disease management: (i) fungal evolutionary diversity (species diversity of forest pathogens and their ecological niches); (ii) pathogen evolution (how forest pathogens become adapted to their hosts); (iii) forest resistance to disease, especially in relation to tree breeding (trade-offs, tolerance, emerging properties in populations); and (iv) the role of hyperparasites and tree microbiota in the regulation of pathogen populations and disease. Finally, we show how knowledge acquired in an evolutionary framework can translate into recommendations for forest pathology in the future, both from an academic and an applied perspective. Although we focus here on forest pathology as the art and science of forest diseases and their control, it should be reminded that forest pathologists more generally contribute to the ecology of parasitism. In this ecosystem perspective, fungal pathogens are no longer “enemies” but key actors of the evolution and ecology of local communities, and more generally of the ecosystem health (e.g., Burdon 1987; Gilbert 2002; Hudson et al. 2006; Ostry and Laflamme 2009).
2 Towards a better knowledge of the diversity, biogeography, and ecology of forest pathogenic fungi and Phytophthora spp.
The emergence of new diseases has underlined the poor current knowledge of microbial evolutionary diversity. Here, we refer not only to the diversity of organisms as taxonomic units but as evolutionary lineages that have diverged and thus present specific distributions and ecological niches (Taylor et al. 2006). In many cases, new diseases of forest trees and wild animals have been caused by introduced organisms, including fungi and oomycetes, which were not even formally described before their invasive behavior (Kupferschmidt 2012). Well-documented examples are forest epidemics caused by Cryphonectria parasitica on American chestnut, the Dutch elm disease caused by Ophiostoma ulmi and Ophiostoma novo-ulmi, the beech bark disease caused by Neonectria faginata, the Port-Orford cedar rot by P. lateralis and sudden oak death by Phytophthora ramorum (Garbelotto 2008; Loo 2009). Indeed, the kingdom Fungi alone has been estimated to include between one to five million species among which only approximately 5% are identified (Blackwell 2011). Similarly, diversity in taxonomic groups such as Phytophthora spp. is probably still largely undescribed (Brasier 2009; Hansen et al. 2011; Hayden et al. 2013; Hüberli et al. 2013; Oh et al. 2013). Understudied areas of Asia, Central America, or Africa are thought to host a huge diversity of undescribed fungi and fungal-like organisms that probably go unnoticed on their native hosts and could potentially be destructive pathogens if introduced outside their native area. (Hyde 2001; Brasier 2009; Schmit and Mueller 2007; Blackwell 2011). The lack of baseline data on fungal diversity and biogeography makes it difficult to say whether an emerging disease is caused by an alien species or a pre-existing pathogen, which has evolved increased virulence or been favored by changing environmental conditions (Jeger and Pautasso 2008; Shaw et al. 2008; Linzer et al. 2009; Shaw and Osborne 2011). Identifying the origin of fungal introductions solely from historical and geographical arguments is rarely feasible because temporal or spatial data retracing the chronology of invasion are often missing, and species descriptions are inaccurate, making it difficult to separate different putative population sources (see for example Kim et al. 2010; Goss et al. 2014). Once the causal organism has been isolated, only a rigorous phylogenetic study associated with epidemiological studies can decipher the causes of emergence among introduction of an exotic pathogen by human-mediated long-distance transport, changes in climatic conditions, and human disturbance to forests ecosystems, in particular, changes in host density, which often interact (Anderson et al. 2004; Desprez-Loustau et al. 2007; Stukenbrock and McDonald 2008; Stenlid et al. 2011; Santini et al. 2013).
The full recognition of fungal and fungal-like diversity has long been hampered by a lack of diagnostic criteria, but the availability of improved molecular techniques now allows the recognition of fungal species consistent with the Evolutionary Species Concept (Taylor et al. 2000). For example, detailed phylogenetical studies have demonstrated that the fungal lineage causing ash dieback was different from the indigenous European species Hymenoscyphus albidus, which is not pathogenic, and that it was conspecific to the Asian species formerly reported as Lambertella albida (Queloz et al. 2011; Husson et al. 2011; Zhao et al. 2012). In many cases, species defined from morphological traits were shown to be pools of genetically isolated lineages that differed significantly in biologically relevant features such as geographic distribution and pathogenicity (Taylor et al. 2000, 2006). Many plant diseases have thus been shown to be caused by a complex of cryptic species (de Wet et al. 2003; Fitt et al. 2006; Crous et al. 2004; Cai et al. 2011; Pérez et al. 2012). In particular, recent studies using DNA sequences have clarified the taxonomic confusion that existed for some important forest pathogens, leading to revisions such as the split of the red band needle blight of Pinus into two distinct species: Dothistroma septosporum and Dothistroma pini (Barnes et al. 2004), the redefinition of the genus Ceratocystis (de Beer et al. 2014), and the description and characterization of the Gibberella fujikuroi complex (Kvas et al. 2009). Another example is oak powdery mildew in Europe, which was shown to be associated with four different cryptic species often coexisting in the same stands: Erysiphe alphitoides, Erysiphe quercicola, Erysiphe hypophylla, and Phyllactinia roboris (Mougou et al. 2008; Mougou-Hamdane et al. 2010). The geographic origin of these species is still hypothetical (Desprez-Loustau et al. 2011), as is the possibility that some of them evolved in sympatry in their region of origin, as a result of evolutionary branching. Such evolutionary branching could explain their temporal niche differentiation, in which certain species predominate at different points in the season (Feau et al. 2012; Hamelin et al. 2011).
The resolution of species complexes and the reappraisal of fungal phylogeny has not only produced taxonomic clarification but also changed our views on the biology of the fungi and their ecological niche. For example, Fabre et al. (2011) showed that the two different Dothistroma species causing similar foliar symptoms in pines, D. septosporum and D. pini, have a different climatic niche, the latter being present in warmer climate. The use of species-specific tools proved that the disease outbreak in the 1990s in France could not be explained by a recent introduction of D. pini, as it was already present in the country in old herbarium samples, but rather by increased temperature conditions in recent years. The long-held belief that powdery mildews, like many biotrophic pathogens, are highly specialized was put into question when the same species were found on distantly related hosts, as were E. alphitoides and E. quercicola on various tropical hosts in plant families very distant to Fagaceae (Takamatsu et al. 2007). Similarly, Fusarium circinatum, part of the large G. fujikuroi complex (Kvas et al. 2009), which had been thought to be specialized on the Pinus genus, was recently shown to be able to behave as an endophyte of different grasses (Swett and Gordon 2012) and to infect maize (Swett and Gordon 2015). This finding raises the question whether grass species can act as reservoir for F. circinatum.
Pathogens that cause minor damage are often unseen—especially in the region of origin of diseases, where pathogens and hosts have coevolved (Parker and Gilbert 2004; Loo 2009; Jousimo et al. 2014). Tracking the origin of invasive fungi and Phytophthora spp. has thus improved our knowledge of these organisms in their natural environments. A few well-documented epidemics caused by introduced pathogens have allowed the reconstruction of the invasion pathways or the exclusion of potential sources by testing different scenarii of introduction, using recent advances in molecular and statistical Bayesian analysis (Grünwald and Goss 2011; Barrès et al. 2012; Dutech et al; 2012; Graça et al. 2013; Tsui et al. 2014). For instance, molecular studies completed since the European invasion of Hymenoscyphus fraxineus strongly suggested that the fungus originates from East Asia, (Gross et al. 2014), where no epidemics have been reported on the local ash species. The number of described Phytophthora species has dramatically increased in the last years, and it is now clear that forest soils host numerous and diverse resident communities of Phytophthora species (Hansen 2008). The bulk of the increase is attributable to better survey and discrimination of Phytophthora spp. in temperate forests and wild land ecosystems (e.g., Burgess et al. 2009; Oh et al. 2013). Whereas these surveys originally focused on highly pathogenic and mostly introduced Phytophthora species, they allowed the discovery of several species unassociated with disease symptoms (Kroon et al. 2011; Hansen et al. 2011). One example is Phytophthora uniformis which does not cause decline of its host, Alnus incana, in its native environment in Alaska (Adams et al. 2009), but is one of the parental species of P. × alni, which has decimated riparian populations of black alder in Europe since the early 1990s (Brasier et al. 1995, see below for the hybridization process).
3 Pathogen evolution: understanding and predicting the success of invasive forest pathogens and the adaptation of pathogens in intensively managed plantations
Invasions by forest pathogens most often lead to new species interactions, without any previous coevolutionary history (Parker and Gilbert 2004). This differs from what is observed in widely planted crop plants, which have a worldwide distribution and are mainly affected by pathogens with which the plant species coevolved in the center of origin. New interactions in these cases occur at the intraspecific level between cultivars and strains of the pathogens, after a period of interruption in the natural coevolutionary process, as in the famous example of potato late blight in Europe (Desprez-Loustau et al. 2007). How a species introduced into a new environment can thrive and develop biotic interactions in the invaded community is a central question in invasion ecology (Elton 1958; Parker and Gilbert 2004; Facon et al. 2006; Keller and Taylor 2008; Vacher et al. 2010). In particular, the outcomes of evolutionary novelty in plant–pathogen interactions, i.e., new encounters between pathogens and naive plants, have led to contradictory predictions. Some have argued for biotic resistance: that is, the novelty of a new environment will be too difficult to overcome, thus only a fraction of introduced pathogens will cause disease (Newcombe and Dugan 2010). Indeed, it has been shown for plants that most biological invasions fail (Williamson and Fitter 1996). In contrast, evolutionary novelty has been suggested to generally benefit plant enemies, since there will have been no selective pressure on resident plants to develop or maintain resistance to the new antagonist (Parker et al. 2006). Verhoeven et al. (2009)) proposed that the outcomes of novel interactions between plants and pathogens or insects can be predicted by the mechanisms underlying the interaction. In particular, novelty is expected to benefit pathogens in the case of recognition-based defense, as occurs in the elicitor–receptor system (Jones and Dangl 2006), since, in this case, the plants have not evolved receptors able to recognize the specific elicitors of the new pathogen and ultimately to trigger resistance responses.
The success of pathogen invasions relies on various processes, from preadaptation or plasticity to postinvasion adaptation (Agosta and Klemens 2008). Deciphering the relative contributions of these processes requires genetic and phenotypic comparisons between source and introduced populations (Keller and Taylor 2008). These analyses have been only rarely performed for forest pathogens, notably in a study demonstrating key differences in transmission between native and introduced species of Heterobasidion (Garbelotto et al. 2010).
In many cases, successful invasive forest pathogens have jumped to new hosts with close phylogenetic relationship, often in the same genus, e.g. C. parasitica from Asian to American or European Castanea spp. (Anagnostakis 1987), Cronartium ribicola from Asian to American or European Pinus spp. (Kim et al. 2010), O. novo-ulmi from Asian to European and American elms (Brasier and Mehrotra 1995), Phytophthora lateralis from Asian to American Chamaecyparis (Brasier et al. 2010), and H. fraxineus from Asian to European ash (Zhao et al. 2012; Goss et al. 2014). The close relationship between original and new hosts fits with the experimental demonstration that the likelihood that a pathogen can infect two plant species decreases continuously with phylogenetic distance (Gilbert and Webb 2007; De Vienne et al. 2009). This relationship between ease of host jumps and phylogenetic distance could be explained by “ecological fitting” or exaptation, i.e., that infectivity traits evolved in a host interaction in its place of origin would allow the pathogen to be able to infect a closely related host species in its area of introduction (Agosta and Klemens 2008; Agosta et al. 2010). A second, not mutually exclusive hypothesis is that fewer evolutionary changes in the pathogen are required to adapt to a new host more closely related to its original host (Altizer et al. 2003; Schulze-Lefert and Panstruga 2011). Only a few mutations events in pathogen effectors and their targeted host proteins were shown to explain successful host jumps among sister species in Phytophthora (Dong et al. 2014). The phylogenetic signal in plant pathogen–host range could help identify potential risks for forest ecosystems and cautions against the introduction of seedlings or trees of exotic species to an area with wild relatives among the indigenous trees (Vacher et al. 2010; Gilbert et al. 2012). However, the generality of these predictions is questioned by the observation of several invasions with apparent host jumps to more distantly related hosts, for example, in the case of P. ramorum (Hansen et al. 2005; Brasier and Webber 2010), oak powdery mildew (Takamatsu et al. 2007; Desprez-Loustau et al. 2011), or Neofusicoccum parvum (Sakalidis et al. 2013). In some instances, ecological proximity plays a larger role than phylogenetic proximity in host jumps (Roy 2001; Refregier et al. 2008). Agosta et al. (2010) proposed a scenario to explain hosts jumps by specialized pathogens with a two-step process. In a first phase, host expansion is favored by ecological perturbations, which put into contact pathogens and new hosts, with a high level of stochasticity. The success of this first phase, i.e., the initiation of a new interaction, is enabled by mechanisms such as ecological fitting or exaptation, which do not require evolutionary innovation. This phase is followed by a phase of host specialization based on evolutionary changes.
This raises the question of the evolutionary potential for introduced pathogens. Indeed, species introductions are often accompanied by severe genetic bottlenecks (Lee 2002). The resulting reduced genetic diversity could happen to be insufficient to allow adaptation to the new environment (i.e., the invasion paradox, Frankham 2005). Multiple introductions from genetically divergent populations have often been invoked to explain the successful invasion of plants or animals despite this apparent paradox (e.g., Kolbe et al. 2004; Lavergne and Molofsky 2007). Multiple introductions not only increase the propagule pressure but also increase overall population genetic variance, hence evolutionary potential. In particular, recombination can occur between previously allopatric, divergent genetic lineages, what is called genetic admixture (Rius and Darling 2014). Such intraspecific hybridization generates novel allelic combinations that can be beneficial for the colonization of new hosts and environments (Lavergne and Molofsky 2007; Rius and Darling 2014). Multiple introductions have been documented for some forest pathogens, such as C. parasitica in Europe (Dutech et al. 2010), F. circinatum in Spain (Berbegal et al. 2013), or Diplodia pinea in South Africa (Burgess et al. 2001). Genetic admixture among genetically divergent populations has been identified in some examples, such as C. parasitica (Dutech et al. 2012), but the demonstration of a benefit for invasion has not yet been rigorously established (Dlugosch and Parker 2008). The respective roles of admixture and adaptation within populations thus represent an important and as-yet unanswered question in understanding the causes of invasive success in forest pathogens.
Interspecific recombination events involving nuclear and mitochondrial genomes have been increasingly recognized as an important factor in the evolution of plant pathogens and their adaptation to new environmental conditions, for example enabling them to perform host jumps between phylogenetically divergent species (Newcombe et al. 2000; Schardl and Craven 2003; Parker and Gilbert 2004; Giraud et al. 2008a,b; Stukenbrock and McDonald 2008; Stukenbrock 2013). Partial sexual compatibility between phylogenetically close species (Le Gac and Giraud 2008) and regular non-Mendelian genetic transmission (Roper et al. 2011) are commonly observed in fungi. These genetic processes increase the probability of genetic introgression among species and, therefore, the likelihood of acquisition of new genes associated with pathogenic functions (Dhillon et al. 2015). Furthermore, the dramatic increase in worldwide movement of pathogens through plant trade (Roy et al. 2014) has favored the encounter in the same environment of phylogenetic taxa that did not develop interfertility barriers during allopatric speciation (Taylor et al. 2006; Le Gac and Giraud 2008). Recent studies on Phytophthora alni, the causal agent of alder decline in Europe, clearly illustrate the consequences of hybridization on the emergence of a new disease. A hybrid origin of the P. alni complex was first concluded from the study of morphological and physiological characters (Brasier et al. 1999). The investigation of ploidy and mitochondrial and nuclear genes in the P. alni complex have confirmed that the epidemic taxon P. xalni was the result of an interspecific hybridization between two close phylogenetic species, P. uniformis and Phytophthora × multiformis (Ioos et al. 2006; Husson et al. 2015). In addition, genetic studies using polymorphic microsatellite markers strongly suggested that the parental species P. uniformis was alien to Europe (Aguayo et al. 2013) and that its introduction subsequently led to its hybridization with P. × multiformis and to the emergence of the aggressive hybrid species P. × alni, which is responsible for the epidemics.
Once the species barrier has been overcome and very high mortality rates have occurred in a new host, it has long been supposed that pathogen populations should evolve towards lesser virulence, in the meaning used by evolutionary biologists, i.e., the amount of damage a parasite causes to its host, encompassing both infectivity (the ability to colonize a host) and severity of the disease (Frank 1996; Parker and Gilbert 2004). Such a decline in virulence is expected as populations move toward equilibrium so long as there are trade-offs between pathogen transmission and virulence—that is, pathogens that cause their hosts to die more quickly have fewer opportunities to spread (May and Anderson 1983; Frank 1993; Alizon et al. 2009). In the case of oak powdery mildew, for example, the severity of epidemics in Europe (associated with mortalities) rapidly declined after the first outbreaks. However, whether this decreased disease severity is attributable to a decreased virulence of the pathogen or to other factors (e.g., changes in forest practices, such as abandonment of pollarding and coppicing, or regulation by hyperparasites) remains unclear (Mougou et al. 2008). In other, long-time invasions, no decrease in virulence has yet been observed (e.g., O. novo-ulmi in Europe or C. parasitica in North America; Jarosz and Davelos 1995). In addition to the classic virulence/transmission trade-off and subsequent intermediate optimum, several factors such as cost of virulence/resistance, trait correlations, and geographic distributions can determine the outcomes for virulence evolution (Bull 1994; Frank 1996; Gandon 2002; May and Anderson 1983). Moreover, the transmission-virulence trade-off may be released in some situations, especially when high transmission rates are favored, allowing virulence to be maintained or increased (Gandon and Michalakis 2002). The strength of the virulence-transmission link varies according to pathogen life-history traits. For example, transmission is expected to remain high even for virulent strains for pathogens that can persist on their dead host as saprobes, as is often observed for necrotroph forest pathogens (e.g., Prospero et al. 2006 for C. parasitica). An increase in virulence is also expected to occur as a consequence of improved pathogen transmission in artificialized ecosystems, such as agroecosystems or planted forests (Stukenbrock and McDonald 2008). Large host plants surfaces with low genetic diversity maintain large pathogen populations, with high evolutionary potential and great opportunities for transmission, thus allowing rapid adaptation to the hosts (Stukenbrock and Bataillon 2012). Increased incidence of qualitative virulence types in response to forest host density has been clearly demonstrated, as in the examples of the breakdown of rust resistance types in plantation poplars (Pinon and Frey 2005). Similarly, C. ribicola populations have been demonstrated to increase in the proportion of strains carrying virulence types in the presence of five-needle pines with corresponding qualitative resistance (Kinloch et al. 2004, 2008). Increased virulence in the quantitative sense (often referred to aggressiveness by plant pathologists ) has been reported in some agroecosystems (Milus et al. 2009; Delmotte et al. 2014). The risk of evolution of increased virulence in response to quantitative resistance has been acknowledged for forest diseases (Ennos 2001), but there is still no clear empirical demonstration. Increasing damage caused by some endemic pathogen species following the extension of host plantations on large surfaces have been reported in the last decades (Woods et al. 2005; Lieberei 2007; Fabre et al. 2011; Xhaard et al. 2012). However, climatic changes (Fabre et al. 2011; Woods et al. 2005) or simply the increase in host density and extent (Lieberei 2007; Xhaard et al. 2012) have been invoked as causes of increasing damage in these studies. Because planted forests are expected to increase rapidly in the next future, special attention toward predicting pathogen evolution in interaction with future forest management practices is surely required.
4 Disease resistance: revisiting the ideotype concept for breeding trees
The forces that allow disease to persist at an overall moderate level in otherwise undisturbed ecosystems have long been a subject of study and speculation (May and Anderson 1983). Wide genetic variation in host resistance and pathogen virulence have been evidenced in such ecosystems, where they are sustained by and result from coevolutionary dynamics between hosts and pathogens (Burdon and Thrall 2000; Keesing et al. 2006; Burdon et al. 2013; Ennos 2015; Jousimo et al. 2014). In contrast, the association between low diversity and high disease incidence has been called the “monoculture effect” in reference to the agricultural context in which frequent, severe epidemics require the use of pesticides to maintain yield loss below acceptable levels (King and Lively 2012). This model of intensification with a strong dependence on external, fossil fuel inputs has been increasingly put into question, and new paradigms for intensification have been widely adopted (Tittonell 2014). However, recent forest intensification has largely followed the old model, with a great expansion of monospecific, and sometimes monoclonal, plantations (Paquette and Messier 2009). The philosophy of some tree breeding strategies still strongly relies on selecting or designing an ideal tree, i.e., “superior genotypes” for mass propagation using biotechnology and genome engineering (Campbell et al. 2003, Harfouche et al. 2012). The questions raised by this approach—what should be an ideal tree? how to select the ideal tree?—have been addressed by many studies and reviews (e.g., Dickmann et al. 1994; Martin et al. 2001; Dubouzet etal. 2013; Franklin et al. 2014). Here, we would like to highlight three topics that have been comparatively little dealt with or applied in the field of tree breeding: trade-offs, tolerance, and group selection.
The idea of an “ideal tree” derives from the ideotype concept, first coined by Donald (1968) for crop plants. In general, targeted phenotypes are mostly defined from growth-related traits (Martin et al. 2001; Harfouche et al. 2012). However, selecting fast-growing trees may be at the expense of defense traits, due to trade-offs between growth and defense (Franklin et al. 2014). A predicted lower resistance to pathogens and herbivores in faster growing plants due to the metabolic cost of resistance has gained empirical support (Lind et al. 2013), and has been observed in trees (Loehle and Namkoong 1987; Fine et al. 2006; Oliva et al 2012, 2014). In a recent study with Populus trichocarpa, McKown et al. (2014) demonstrated the occurrence of trade-offs in the evolution of stomatal traits that were positively related to growth (photosynthesis and shoot elongation) and negatively to defense (foliar tannins and Melampsora infection). Such negative relationships between growth and disease resistance could have important implications for breeding programs. A recent meta-analysis demonstrated that the selection of poultry lines for rapid growth unintentionally resulted in a strongly reduced immune function (Van der Most et al. 2011). The authors however concluded that, by explicitly taking the relative costs of growth and immune functions into consideration during selection, it may be possible to breed animals for increased growth without loss of immune function. It has also been shown for plants that the growth-defense trade-offs are complex and may vary according to the resistance traits involved, e.g., constitutive vs. induced resistance, or resistance sensu stricto vs. tolerance (Kempel et al. 2011). Moreover, environmental constraints also interact with the growth-defense trade-off to determine fitness. In a recent study, Franklin et al. (2014) proposed a mechanistic model of tree fitness integrating the most relevant traits involved in growth and survival (including defense allocation), in addition to limiting factors, such as light environment and drought, disturbance frequency (including pests and pathogens), and competition. This modeling framework can be used to explore the potential and limitations of artificial selection to increase productivity in a managed forest environment, by identifying optimal combinations of traits for a given goal and explicitly relating expected productivity gains to changes in tree vulnerability to biotic and abiotic risks in various environmental conditions. Using a similar framework, selection for qualitative disease resistance in limber pine has been shown to alter trees’ drought and temperature tolerances, potentially changing their environmental range (Vogan and Schoettle 2015).
When dealing with plant (tree) defense against pathogens, much emphasis has been put on resistance in its restricted meaning, i.e., mechanisms that prevent or limit the development of the pathogen (see review in Telford et al. 2015). These mechanisms therefore exert a selection pressure on pathogen populations, the strength of which depends, on the plant side, on the level of resistance (from partial to complete) and the spatial scale of the deployment of resistant genotypes (McDonald and Linde 2002; REX Consortium 2013; Ennos 2015). The evolutionary constraints affecting the long-term efficacy of resistance strategies are well illustrated by past failures in the breeding programs of rust resistant poplar cultivars in Europe. The complete resistance conferred by race-specific genes widely deployed in monoclonal plantations was rapidly overcome by virulent races of the pathogen, in several boom-and-bust cycles (Pinon and Frey 2005). These failures have pushed breeders to consider other types of resistance, such as polygenic partial resistance, with the goal to slow down the evolution of virulent pathogen variants, thus increase the durability of resistance (Dowkiw et al. 2012). The increasing knowledge of plant–pathogen molecular interactions and technological advances are enabling the discovery and functional profiling of new resistance genes, the breakdown of which would incur high fitness costs in corresponding virulent pathogens (Vleeshouwers et al. 2008; Michelmore et al. 2013). Tolerance in its restricted meaning (i.e., mechanisms that reduce the host fitness costs of infection, Roy and Kirchner 2000; Simms 2000) has been the subject of a growing interest as a potentially more stable defense component than true resistance since it was considered to place little or no selective pressure on the pathogen (Schafer 1971; Ney et al. 2013; Ennos 2015). Tolerance, however, is in nearly all cases expected to impose positive feedbacks leading to more prevalent and/or more virulent pathogens (Little et al. 2010; Miller, White, and Boots 2006; Roy and Kirchner 2000). Therefore, introducing tolerant hosts to a population, through breeding for example, has the potential to increase pathogen pressure overall (Restif and Koella 2004). More pathogens in the landscape can lead to negative effects on nontarget host species such as wild relatives (Power and Mitchell 2004), or even to host jumps to new species, since the probability of eventual emergence on new hosts is increased with a larger pathogen reservoir (Antia et al. 2003). While it has been observed that asymptomatic infections can be important drivers in the epidemiology of forest diseases (Elegbede et al. 2010; Denman et al. 2008), the potential for host tolerance to increase disease prevalence in adjacent populations or nontolerant host species is rarely acknowledged in a management context (Hayden et al. 2013; Little et al. 2010). Breeding strategies combining both resistance and tolerance mechanisms therefore appear to be the most desirable (Walters et al. 2012). This requires an accurate assessment of resistance and tolerance traits and their interactions (Simms and Triplett 1994), as investigated in the Douglas fir–Armillaria ostoyae pathosystem (Cruikshank and Jaquish 2014).
Just as disease tolerance has the potential to increase disease overall, a major challenge for breeders lies in the fact that characters of interest are relevant at stand level, while selection is based on individual performance. This issue was central to Colin Donald’s ideotype concept, which balanced individual performance against competitive ability and population performance, but it was largely overlooked (Zhang et al. 1999). As an agronomist, Donald focused on yield, but his arguments are particularly relevant for disease resistance. Disease resistance can be visualized as an emerging property of populations, in that it is strongly affected by density- and frequency-dependent ecological and evolutionary processes (Burdon et al. 2014; Ennos 2015; Telford et al. 2015; Papaïx et al. 2015). Indeed, durable resistance to pathogens is a population attribute that can only be assigned retrospectively (Johnson 1984). Rather than relying on “durable resistance genes,” durable resistance should be considered in terms of a strategy to minimise the pace of evolution of virulence in the pathogen population (REX Consortium 2013; Michelmore et al. 2013). This can be achieved by a variety of approaches based on an adequate deployment of multiple resistance genes either by pyramiding and/or by heterogeneous deployment in space and time, so as to diversify the selection pressure on the pathogen (McDonald and Linde 2002; Michelmore et al. 2013; REX Consortium 2013). Growing clonal mixtures of 5–20 genotypes for short-rotation coppice of willow has proven to be feasible and effective for both disease reduction and yield increase over a long term (McCracken et al. 2011). Nevertheless, such experiments with trees are relatively few, and are centered on genotypes that were selected on their individual performance. Going still further, breeding for genetically diverse populations has received increasing attention in recent years, especially in the context of a “Darwinian agriculture” to address future challenges of changing climate and sustainable management (e.g., Denison et al. 2003; Dawson and Goldringer 2011). In particular, human-mediated (artificial) group selection has been suggested to offer a wider scope for improvement than selection on individual fitness traits (Zhang et al. 1999; Denison et al. 2003). Natural selection is thought to operate mostly among individuals and more weakly at a group level, and is thus expected to have favored traits linked to individual performance even if costly to group productivity. As a consequence, individual traits that improve group performance could offer greater opportunities for artificial selection (Carroll et al. 2014). An application of this concept for resistance traits was recently proposed by Anche et al. (2014), who investigated the use the basic reproduction ratio, R0, which is a key epidemiological parameter defined at the population level, in breeding programs using individual assessments. The theoretical basis of breeding strategies targeting population traits, i.e., an association of “cooperative phenotypes,” has been developed and applied with success in animals and cereals (Ellen et al. 2014; Carroll et al. 2014). Recently initiated tree species diversity experiments have allowed for the testing of associational effects (those linked to interactions between species) on traits including disease resistance and susceptibility (Castagneyrol et al. 2014; Hantsch et al. 2013, 2014). The same type of experiments could be used to explore within-species associational effects, which may be even higher in magnitude than between-species effects (Crawford and Rudgers 2013).
5 Hyperparasitism of fungal pathogens: a poorly understood but promising means of bio-control
The long-stable coexistence of plants and their pathogens is generally explained by coevolutionary processes (Newton et al. 2010; Jousimo et al. 2014). However, pathogens and plants are included in larger networks where interacting species may also contribute to the natural regulation of disease (Lafferty et al. 2006; Selakovic et al. 2014). In particular, trophic cascades, where negative effects of herbivores on plant biomass are reversed by the effects of herbivore predators, have been widely studied and used in agricultural and forest entomology systems, including predatory insects, parasitoids, or disease-causing pathogens (Wainhouse 2005). Developing these methods for pathogens presents a challenge to forest pathologists and managers, in order to face increasing forest disease risks in the context of global change and new plantations. A necessary prerequisite for achieving this goal is improved understanding of mycoparasite diversity, ecology, and evolution (Roderick et al. 2012).
Because of their deleterious effect on host fitness (i.e., virulence), hyperparasites play a key role in the ecology and the evolution of parasite populations (Tollenaere et al. 2014). Koskella (2013, 2014) has nicely demonstrated that bacterial populations on horse-chestnut trees are engaged in coevolutionary dynamics with their virus phage parasites and moreover exhibit reciprocal adaptation across time and space. However, very few tri-trophic interactions involving a mycoparasite, a fungal or oomycete pathogen, and a tree host have been studied so far. This can be explained by the difficulty of detecting the hyperparasites (the biodiversity of which is even less known that that of parasites) and the complexity of these interactions due to the life cycles of both microbial protagonists. The best studied examples of hyperparasites of fungi are hypovirulent viruses, which decrease the virulence of their fungal hosts [e.g., Cryphonectria Hypovirus1 (CHV1) infecting the chestnut blight fungus and Dutch elm fungi O. ulmi and O. novo-ulmi]. Their role in the regulation of diseases has been established (Davelos and Jarosz 2004; Springer et al 2013; Swinton and Gilligan 1999). Insights gained in CHV1 ecology (phenotypic and genetic diversity, transmission and evolution), which is used for biocontrol in chestnut orchards (Grente 1981) could help to improve biocontrol strategies. However, fungi can also be parasitized by other fungi (Barnett 1963; Kiss 2003). This fungal lifestyle has attracted an increased interest in recent years, with studies of the ecological impact of mycoparasites on fungal host populations (Tollenaere et al. 2014) and investigations about their mode of action at the molecular level (Mukherjee et al. 2013; Siozios et al. 2015). Metagenomic studies applied to mycoviruses (Pearson et al. 2009; Ghabrial and Suzuki 2009; Roossinck et al. 2010; Xie and Jiang 2014) and fungi (see next section) point to new hyperparasite or fungal antagonist candidates. Some mycoviruses have followed the route of invasion of their invasive fungal hosts and have been first detected in the introduced range of their hosts (Bryner et al. 2012; Feau et al. 2014; Voth et al. 2006; Schoebel et al. 2014; Martinez-Alvarez et al. 2014). However, according to the enemy release hypothesis (Mitchell and Power 2003), hyperparasites are expected be more frequent in the native range of their hosts than elsewhere, and thus, systematic searches should be concentrated in these areas (Roderick and Navajas 2003; Hale et al. 2014; Ridout and Newcombe 2015). Such a search for hyperparasites requires a known centre of origin of the parasite, which is far from the rule, as seen before.
Releases of biocontrol agents outside their native ranges have not always been successful (Roderick and Navajas 2003). For example, in spite of several introductions of CHV1 in Northern America, the hyperparasite never established there (Milgroom and Cortesi 2004). As it has been shown for invasive species, hyperparasite establishment success depends on demographic, genetic, and environmental factors (Hufbauer and Roderick 2005; Roderick et al. 2012; Fauvergue et al. 2012). The identification and selection of transmission and virulence traits is a necessary step for short-term success of hyperparasites strains for biological control (Robin et al. 2010; Swinton and Gilligan 1999). For different reasons, transmission and virulence are not easy to quantify for hyperparasites. Most mycoviruses have no extracellular life stages and are transmitted both vertically and horizontally (Pearson et al. 2009). The horizontal transmission of mycoviruses is controlled by the fungal vegetative incompatibility system, which, until recently, was thought to impede virus transmission in populations. However, some mycoviruses have a global distribution [e.g., CHV1 (Bryner et al 2012), Gremmenellia virus (Botella et al 2014)]. Recent results showed that in situ transmission rates of CHV1 are higher than in vitro studies had suggested (Carbone et al. 2004; Brusini and Robin 2013). Moreover, different strategies, aiming at decreasing the vegetative incompatibility reaction, finding a universal mycovirus donor, or creating vectors for mycoviruses, have been proposed to increase transmission efficiency of mycovirus within and among fungal species (Xie and Jiang 2014). Mycovirus virulence results from interactions of both parasite and hyperparasite genomes, which give it the attribute of an extended phenotype (Lambrechts et al. 2006). In conditions where transmission between hosts is not restricted, “mild” hyperparasite strains, characterized by low virulence and high transmission rates, are more apt to regulate disease than severe strains because they more readily establish in a host population than severe strains (Morozov et al. 2007). When between-host transmission is restricted by fungal vegetative incompatibility (transmission rates varying from 0 to 100%), mycoviruses exert a frequency-dependent selection on their host, which increases with increasing virulence of the mycovirus, resulting in an increase in fungal population diversity (Brusini et al. 2011). However, highest levels of polymorphism in the host population result in unfavorable conditions for mycovirus transmission. Thus, by this feedback mechanism, less virulent mycoviruses are likely favored in diverse fungal host populations. This theoretical result supports regulation efforts to impede an increase in the diversity of European C. parasitica populations in Europe where biological control of chestnut blight is still effective and used.
Establishment success of hyperparasites may also be strongly dependent on the biotic and abiotic environment. Milgroom and Cortesi (2004) have argued that failures of biological control against chestnut blight disease in eastern North America could be linked to forest ecosystem characteristics, quite different from the European orchard agroecosystems where virus-infected strains are successfully released. This suggests that biological control may have greater success in forest plantations with higher tree density and lower heterogeneity than in seminatural forests, due to the facilitated development of the fungal parasite (Xie and Jiang 2014). Moreover, virus × fungus × environment interactions also affect the outcome of hyperparasite × parasite interactions (Bryner and Rigling 2011), which suggests that adaptation of mycovirus lineages to new environments can occur.
The hyperparasite’s evolutionary history can also provide useful insights to develop a successful biocontrol strategy. Genetic analyses of CHV1 populations in western and southern Europe have demonstrated that establishment and spread of the mycovirus are associated with a few recombinant virus lineages (Feau et al. 2014). These results, together with the known effect of hybridization on vigor of biological control agents (Szucs et al. 2012), the high rate of beneficial recombinations in viruses (Vuillaume et al. 2011), and the demonstrated high cost of bacterial resistance in coevolution with multiple phage viruses (Koskella et al. 2012) suggest to use several hyperparasite lineages in biological control strategies, especially in areas where releases of hyperparasites have failed until now. Coinfection strategies should be thoroughly studied in experimental systems prior to deployment, since the coexistence of different strains of a mycovirus within one host may not only lead to homologous recombinations but also result in the selection of more virulent strains, with lower transmission efficiency (Frank 1996; Mosquera and Adler 1998).
6 The tree microbiota: a rapidly evolving trait that will allow trees to face anthropogenic environmental change?
Due to their short generation time, pathogens are expected to quickly adapt to human-driven environmental change through changes in distribution and physiology, with or without host switches. By comparison, trees are expected to adapt more slowly and to experience a phenotype–environment mismatch (Carroll et al. 2014). There is, however, one facet of the tree phenotype that may evolve as quickly as the pathogens: the tree microbiota, which belongs to the extended phenotype of a tree (Dawkins 1999; Whitham et al. 2006; Partida-Martínez and Heil 2011). Indeed, virtually, all tissues of trees are colonized by microorganisms (Turner et al. 2013). These microbial communities, which are the outcome of millions of years of coevolution (Partida-Martínez and Heil 2011), modulate the tree’s resistance against multiple stresses, including pathogens (Arnold et al. 2003; Hacquard and Schadt 2015). Within the current context of rapid environmental change, it is thus crucial to tackle the following issues: (i) Which properties of the resident microbiota protect the tree against biotic and abiotic stresses? (ii) How will these properties respond to environmental change? (iii) Can we select tree genotypes with beneficial microbiota to facilitate their adaptation to environmental change? Theoretical ecology provides a powerful framework for approaching the first issue. For instance, the biodiversity–ecosystem function (BEF) theory posits that biodiversity increases the intensity and stability of ecosystem functions (Cardinale et al. 2012). Future research aimed at improving tree performance could examine whether this relationship holds at the individual level. For instance, it could examine whether the diversity of tree microbiota improves the various components of tree fitness, including the resistance to pathogens. Such microbiota diversity-individual fitness (MIF) relationship can now be studied by using new sequencing technologies (Di Bella et al. 2013; Vayssier-Taussat et al. 2014), which allow the various properties of tree microbiota (community richness; taxonomic, phylogenetic, or functional diversity; and composition) to be readily described (Buée et al. 2009; Jumpponen and Jones 2009). By combining these novel technologies with methods for network inference (Faust and Raes 2012), the networks of microbial interactions can also be deciphered. Such an approach could enable the identification of potential pathogen antagonists, and thus facilitate protection strategies based on microbial epiphytes and endophytes (Witzell et al. 2014). This approach may also reveal network properties driving the stability of the residential microbiota, in particular its invasibility by pathogens (Kemen 2014).
The second issue is to assess how fast and in which direction the beneficial properties of the resident microbiota will respond to environmental change. There is already a large body of knowledge showing that the composition and structure of the tree microbiota strongly respond to environmental factors such as temperature, drought, atmospheric CO2, or urbanization (Jumpponen and Jones 2010; Compant et al. 2010; Peñuelas et al. 2012; Cordier et al. 2012a; Coince et al. 2014; Rico et al. 2014). There is also increasing evidence showing that the tree microbiota is influenced by the genetic variability of trees, both at intraspecific (Schweitzer et al. 2008; Cordier et al. 2012b; Bálint et al. 2013) and interspecific (Knief et al. 2010; Redford et al. 2010; Kembel and Mueller 2014; Kembel et al. 2014) levels. Tree microbiota properties can thus be considered to be phenotypic traits that are influenced by both the environment and the tree genotype (Bálint et al. 2015). Phenotypic plasticity may induce a rapid change in tree microbiota properties, in response to anthropogenic environmental change. This change may then be followed by an evolutionary change, caused by the genetic adaptation of tree populations (Donnelly et al. 2012). Both mechanisms, plasticity and genetic adaptation of the tree microbiota, may allow tree populations to cope with novel, emerging pathogens; thus, a current challenge for forest pathologists is to assess their relative influence.
Finally, the last challenge will be to integrate the beneficial properties of the tree microbiota into the selection process for tree genotypes (Newton et al. 2010; Chakraborty and Newton 2011). After the characterization of the beneficial properties through observations and manipulative experiments, their genetic architecture will have to be deciphered and their heritability to be assessed (Peiffer et al. 2013; Horton et al. 2014). Evolutionary models simulating the complex network of interactions formed by the novel tree genotypes, their pathogens, and microbiota (Kemen 2014) could then be used to define tree breeding strategies that will sustain forest health. However, such simulations are far from being trivial. Numerous evolutionary models have been developed in the case of simple, pairwise plant–pathogen interactions (Gilligan 2008; REX Consortium 2013). Extending these models to a greater number of species remains a challenge for evolutionary biologists (Brännström et al. 2012; Fortuna et al. 2013).
7 Conclusions and recommendations for the future
At the turn of the twenty-first century, Paul Manion (2003) reviewed some recently evolved concepts in forest pathology, which had modified the way of thinking about diseases and their practical management. Shortly more than 10 years after, the ecological view advocated in this prospective article has gained increasing recognition, and has also been the subject of substantial new evolution. It is remarkable to note that neither of the terms “global” nor “evolution” (except for that of concepts) were used by Manion. The recognition of global change in all its dimensions, including not only climate change but also accelerating global trade, habitat destruction, and other human-caused effects on the environment, has profoundly affected all areas of science and society in the last 15 years. In particular, two major changes affecting forest pathology—the world movement of species with trade, and the rise of plantation forestry to meet growing needs of an increasing human population—have led to an increasing number of emerging diseases, mobilizing the efforts of forest pathologists, and these trends are expected to continue (Essl et al. 2011; FAO 2010). In this context of global changes, characterized by strong new selective pressures put on entire ecosystems by human activities, the coupling of ecology and evolutionary biology has become essential for the understanding of forest diseases and their management. Three important advances, which have important practical implications and which also open new questions for future research, can be highlighted (see Table 1).
-
1.
Not everything is everywhere (especially forest pathogens)
The view on the processes underlying geographic variation in microbial distributions has changed from purely ecological explanations (“everything is everywhere but the environment selects,” according to Beijerinck’s and Becking’s famous claim; see in O'Malley 2008) to a combination of ecological and evolutionary (diversification) explanations supported by evidence of limited (natural) dispersal in many microbial taxa (Taylor et al. 2006; Hanson et al. 2012; Tedersoo et al. 2014 ). Human-mediated transfers putting into contact non-coevolved species therefore result in ecological and evolutionary dynamics that challenge processes operating in natural communities (Burdon et al 2013). These new introductions may overwhelm the ability of the community, or even of artificial selection, to respond (Cobb et al. 2012). For instance, European and American chestnuts have been successively impacted by three exotic pests and pathogens in the last century, Phytophthora cinnamomi, C. parasitica, and Dryocosmus kiruphilus, showing the increasing difficulty for breeding programs to keep pace with biotic threats in woody perennials (Anagnostakis 2012).
-
2.
Pathogen evolution can occur at short time scales
The recognition that evolution can take place at short time scales is not very new (e.g., Cook and Askew 1970), but it has been increasingly supported by evidence linked to anthropogenic changes, such as breakdowns of human-selected resistance in a medical or agricultural context (McDonald and Linde 2002; REX 2013) and adaptation of invasive organisms (Gladieux et al. 2015). Forests have already paid heavy tribute to invasions by exotic pathogens, but still, limited domestication of forest trees has up to now restricted impacts on forest pathogen virulence as those seen in agriculture. Phytosanitary problems in monospecific (monoclonal) planted forests are likely to increase in the future, since experience demonstrates that high productivity achieved in the first rotations, often associated with enemy escape, is likely to fade away with time due to the progressive arrival of natural enemies, i.e. tree pests and diseases (Harwood and Nambiar 2014; Liebhold and Wingfield 2014). New prospects for an ecological and evolutionary intensification could benefit from rapid changes in technology and wood usages as material or fuel (e.g., engineered wood products, composite materials, wood pellets), which will likely remove some of the obstacles to the use of diverse plantations.
-
3.
The tree is a multitrophic community
Community fingerprinting methods, and, more recently, high-throughput sequencing techniques, have revealed that the epiphytic and endophytic microbial communities are much more complex than previously realized (e.g., Yang et al. 2001; Jumpponen and Jones 2009). Pathogens themselves host a diversity of parasites, especially mycoviruses ( Witzell et al. 2014). Some experimental manipulations at small spatial scales have highlighted the role of microbial communities in disease control (Arnold et al. 2003; Matos et al. 2005), while observations at larger scales have revealed environmental factors driving these communities (Cordier et al. 2012a; Peñuelas et al. 2012). Both approaches are required to predict, and ultimately manage, microbial interactions and functions in the context of global change (Kinkel et al. 2011; Berlec 2012; Borer et al. 2013; Lebeis 2014; Vayssier-Taussat et al. 2014; Hacquard and Schadt 2015). Future studies working toward this aim will benefit from the development of functional metagenomics (Knief et al. 2012), microbial interactions modeling (Faust and Raes 2012), as well as evolutionary platforms predicting the outcomes of multispecies interactions (Fortuna et al. 2013).
Uncertainties, especially regarding the emergence of new diseases, are likely a key component of the future of forest pathology (Shaw and Osborne 2011). An obvious policy recommendation is therefore to maintain the expertise in the taxonomy, biology, epidemiology, and ecology of forest fungal pathogens needed to respond quickly to disease outbreaks in more economically developed countries (BSPP 2012). Special efforts should also be developed in regions, such as tropical areas, where high biological diversity is present but has received limited attention so far due to a lack of funding and trained researchers (Kamgan et al. 2008). Spectacular advances and reducing costs in DNA technologies have recently enabled considerable progress in the knowledge of phylogenetic diversity (inter- and intraspecies) of forest pathogens. However, an ecological and evolutionary forest pathology cannot neglect the study of phenotypes and functional traits. Investigations of a greater number of forest pathosystems (not only focused on recent introductions) and the development of high-throughput phenotyping methods are certainly deserving more efforts.
Forest pathologists have increasingly tried to disseminate their research outcomes to contribute to policy recommendations (e.g. the Montesclaros declaration, http://www.iufro.org/science/divisions/division-7/70000/publications/montesclaros-declaration/; Cavers 2015). Increasing links between biologists, sociologists, and economists, for example to investigate the perception of risks or the economic impacts of pathogens, or to assess the ecological service of disease regulation, could help identify the best levers to tackle forest health issues in a globalised and changing environment (Stenlid et al. 2011; Fears et al. 2014). Particular effort should also be dedicated to education, not only to raise awareness in the public and stakeholders of forest health issues and their dependence on human activities but also to attract young scientists, from diverse scientific horizons, to the field of forest pathology.
References
Adams GC, Catal M, Trummer LM (2009) Distribution and severity of Alder Phytophthora in Alaska. In: Proceedings of the sudden oak death fourth science symposium (ed. by Gen. Tech. Rep. PSW-GTR-229. Department of Agriculture FS), pp. 29–49
Agosta SJ, Klemens JA (2008) Ecological fitting by phenotypically flexible genotypes: implications for species associations, community assembly, and evolution. Ecol Lett 11:1123–1134
Agosta SJ, Janz N, Brooks DB (2010) How specialists can be generalists: resolving the “parasite paradox” and implications for emerging infectious disease. Zoologia 27:151–162
Aguayo J, Adams GC, Halkett F, Catal M, Husson C, Nagy ZA, Hansen EM, Marcais B, Frey P (2013) Strong genetic differentiation between North American and European populations of Phytophthora alni subsp. uniformis. Phytopathology 10:190–199
Alizon S, Hurford A, Mideo N, Van Baalen M (2009) Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J Evol Biol 22:245–259
Altizer S, Harvell CD, Friedle E (2003) Rapid evolutionary dynamics and disease threats to biodiversity. Trends Ecol Evol 18:589–596
Anagnostakis SL (1987) Chestnut blight: the classical problem of an introduced pathogen. Mycologia 79:23–37
Anagnostakis SL (2012) Chestnut breeding in the United States for disease and insect resistance. Plant Dis 96:1392–1403
Anche MT, de Jong MCM, Bijma P (2014) On the definition and utilization of heritable variation among hosts in reproduction ratio R0 for infectious diseases. Heredity 113:364–374
Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR, Daszak P (2004) Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol Evol 19:535–544
Antia R, Regoes RR, Koella JC, Bergstrom CT (2003) The role of evolution in the emergence of infectious diseases. Nature 426:658–661
Arnold AE, Mejía LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre EA (2003) Fungal endophytes limit pathogen damage in a tropical tree. Proc Natl Acad Sci U S A 100:15649–15654
Bálint M, Tiffin P, Hallström B, O'Hara RB, Olson MS, Fankhauser JD, Piepenbring M, Schmitt I (2013) Host genotype shapes the foliar fungal microbiome of balsam poplar (Populus balsamifera). PLoS One 8, e53987. doi:10.1371/journal.pone.0053987
Bálint M, Barta L, Hara RBO, Olson MS, Otte J, Pfenninger M, Robertson AL, Tiffin P, Schmitt I (2015) Relocation, high-latitude warming and host genetic identity shape the foliar fungal microbiome of poplars. Mol Ecol. doi:10.1111/mec.13018
Barnes I, Crous PW, Wingfield BD, Wingfield MJ (2004) Multigene phylogenies reveal that red band needle blight of Pinus is caused by two distinct species of Dothistroma, D-septosporum and D-pini. Stud Mycol 50:551–565
Barnett HL (1963) The nature of mycoparasitism by fungi. Annu Rev Microbiol 17:1–14
Barrès B, Carlier J, Seguin M, Fenouillet C, Cilas C, Ravigne V (2012) Understanding the recent colonization history of a plant pathogenic fungus using population genetic tools and Approximate Bayesian Computation. Heredity (Edinb) 109:269–279
Bazin E, Mathe-Hubert H, Facon B, Carlier J, Ravigne V (2014) The effect of mating system on invasiveness: some genetic load may be advantageous when invading new environments. Biol Invasions 16:875–886
Berbegal M, Perez-Sierra A, Armengol J, Grünwald NJ (2013) Evidence for multiple introductions and clonality in Spanish populations of Fusarium circinatum. Phytopathology 103:851–861
Berendsen RL, Pieterse CMJ, Bakker P (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486
Bergot M, Cloppet E, Perarnaud V, Déqué M, Marcais B, Desprez-Loustau ML (2004) Simulation of potential range expansion of oak disease caused by Phytophthora cinnamomi under climate change. Glob Chang Biol 10:1–14
Berlec A (2012) Novel techniques and findings in the study of plant microbiota: search for plant probiotics. Plant Sci Int J Exp Plant Biol 193–194:96–102
Blackwell M (2011) The Fungi: 1, 2, 3 … 5.1 million species? Am J Bot 98:426–438
Booth TH, Jovanovic T, Old KM, Dudzinski MJ (2000) Climatic mapping to identify high-risk areas for Cylindrocladium quinqueseptatum leaf blight on eucalypts in mainland South East Asia and around the world. Environ Pollut 108:365–372
Borer ET, Kinkel LL, May G, Seabloom EW (2013) The world within: quantifying the determinants and outcomes of a host’s microbiome. Basic Appl Ecol 14:533–539
Botella L, Tuomivirta TT, Vervuurt S, Diez JJ, Hantula J (2014) Occurrence of two different species of mitoviruses in the European race of Gremmeniella abietina var. abietina, both hosted by the genetically unique Spanish population. Fungal Biol 116:872–882
Brännström Å, Johansson J, Loeuille N, Kristensen N, Troost T, Hille Ris Lambers R, Dieckmann U (2012) Modelling the ecology and evolution of communities : a review of past achievements, current efforts, and future promises. Evol Ecol Res 14:601–625
Brasier CM (1996) Phytophthora cinnamomi and oak decline in southern Europe. Environmental constraints including climate change. Ann Sci For 53:347–358
Brasier CM (2001) Rapid evolution of introduced plant pathogens via interspecific hybridization. Bioscience 51:123–133
Brasier CM (2008) The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol 57:792–808
Brasier CM (2009) Phytophthora biodiversity: how many Phytophthora species are there? General Technical Report—Pacific Southwest Research Station, USDA Forest Service
Brasier CM, Mehrotra MD (1995) Ophiostoma himal-ulmi sp-nov, a new species of Dutch elm disease fungus endemic to the Himalayas. Mycol Res 99:205–215
Brasier CM, Webber J (2010) Plant pathology. Sudden larch death. Nature 466:824–825
Brasier CM, Rose J, Gibbs JN (1995) An unusual Phytophthora associated with widespread alder mortality in Britain. Plant Pathol 44:999–1007
Brasier CM, Cooke DEL, Duncan JM (1999) Origin of a new Phytophthora pathogen through interspecific hybridization. Proc Natl Acad Sci U S A 96:5878–5883
Brasier CM, Vettraino AM, Chang TT, Vannini A (2010) Phytophthora lateralis discovered in an old growth Chamaecyparis forest in Taiwan. Plant Pathol 59:595–603
Brusini J, Robin C (2013) Mycovirus transmission revisited by in situ pairings of vegetatively incompatible isolates of Cryphonectria parasitica. J Virol Methods 187:435–442
Brusini J, Robin C, Franc A (2011) Parasitism and maintenance of diversity in fungal vegetative incompatibility system: the role of selection by deleterious cytoplasmic elements. Ecol Lett 14:444–452
Bryner SF, Rigling D (2011) Temperature-dependent genotype-by-henotype interaction between a pathogenic fungus and its hyperparasitic virus. Am Nat 177:65–74
Bryner SF, Rigling D, Brunner PC (2012) Invasion history and demographic pattern of Cryphonectria hypovirus 1 across European populations of the chestnut blight fungus. Ecol Evol 2:3227–3241
BSPP (2012) Plant pathology education and training in the UK: an audit. http://www.bspp.org.uk
Buée M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S, Martin F (2009) 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol 184:449–456
Bull J (1994) Perspective: virulence. Evolution 48:1423–1437
Burdon JJ (1987) Diseases and plant population biology. Cambridge University Press, Cambridge
Burdon JJ, Thrall PH (2008) Pathogen evolution across the agro-ecological interface: implications for disease management. Evol Appl 1:57–65
Burdon JJ, Thrall PH (2000) Coevolution at multiple spatial scales - from population to continent and beyond. Evol Ecol 14:261-281
Burdon JJ, Thrall PH, Ericson L (2013) Genes, Communities and Invasive Species: Understanding the Ecological and Evolutionary Dynamics of Host–Pathogen Interactions. Curr Opin Plant Biol 16: 1–6
Burdon JJ, Barrett LG, Rebetzke G, Thrall PH (2014) Guiding deployment of resistance in cereals using evolutionary principles. Evol Appl 7:609–624
Burgess T, Wingfield BD, Wingfield MJ (2001) Comparison of genotypic diversity in native and introduced populations of Sphaeropsis sapinea isolated from Pinus radiata. Mycol Res 105:1331–1339
Burgess TI, Webster JL, Ciampini JA, White D, Hardy GES, Stukely MJC (2009) Re-evaluation of Phytophthora species isolated during 30 years of vegetation health surveys in Western Australia using molecular techniques. Plant Dis 93:215–223
Cai L, Giraud T, Zhang N, Begerow D, Cai G, Shivas R (2011) The evolution of species concepts and species recognition criteria in plant pathogenic fungi. Fungal Divers 50:121–133
Campbell MM, Brunner AM, Jones HM, Strauss SH (2003) Forestry's fertile crescent: the application of biotechnology to forest trees. Plant Biotechnol J 1:141–154
Carbone I, Liu YC, Hillman BI, Milgroom MG (2004) Recombination and migration of Cryphonectria hypovirus 1 as inferred from gene genealogies and the coalescent. Genetics 166:1611–1629
Cardinale BJ, Duffy JE, Gonzalez A, Hooper DU, Perrings C, Venail P, Narwani A, Mace GM, Tilman D, Wardle DA, Kinzig AP, Daily GC, Loreau M, Grace JB, Larigauderie A, Srivastava D, Naeem S (2012) Biodiversity loss and its impact on humanity. Nature 486:59–67
Carroll SP, Jørgensen PS, Kinnison MT, Bergstrom CT, Denison RF, Gluckman P, Smith TB, Strauss SY, Tabashnik BE (2014) Applying evolutionary biology to address global challenges. Science 80:1–16
Castagneyrol B, Jactel H, Vacher C, Brockerhoff EG, Koricheva J (2014) Effects of plant phylogenetic diversity on herbivory depend on herbivore specialization. J Appl Ecol 51:134–141
Cavers S (2015) Evolution, ecology and tree health: finding ways to prepare Britain’s forests for future threats. Forestry 88:1–2
Cavers S, Cottrell JE (2015) The basis of resilience in forest tree species and its use in adaptive forest management in Britain. Forestry 88:13–26
Chakraborty S, Newton AC (2011) Climate change, plant diseases and food security: an overview. Plant Pathol 60:2–14
Cobb RC, Filipe JAN, Meentemeyer RK, Gilligan CA, Rizzo DM (2012) Ecosystem transformation by emerging infectious disease: loss of large tanoak from California forests. J Ecol 100:712–722
Coince A, Cordier T, Lengellé J, Defossez E, Vacher C, Robin C, Buée M, Marçais B (2014) Leaf and root-associated fungal assemblages do not follow similar elevational diversity patterns. PLoS One 9, e100668
Combes C (2001) Parasitism : ecology and evolution of intimate interactions. Press, Chicago University, 552 pp
Compant S, van der Heijden MG, Sessitsch A (2010) Climate change effects on beneficial plant-microorganism interactions. FEMS Microbiol Ecol 73:197–214
Cook LM, Askew RR (1970) Increasing frequency of the typical formof the peppered moth in Manchester. Nature 227:1155
Cordier T, Robin C, Capdevielle X, Fabreguettes O, Desprez-Loustau ML, Vacher C (2012a) The composition of phyllosphere fungal assemblages of European beech (Fagus sylvatica) varies significantly along an elevation gradient. New Phytol 196:510–519
Cordier T, Robin C, Capdevielle X, Desprez-Loustau M-L, Vacher C (2012b) Spatial variability of phyllosphere fungal assemblages: genetic distance predominates over geographic distance in a European beech stand (Fagus sylvatica). Fungal Ecol 5:509–520
Crawford KM, Rudgers JA (2013) Genetic diversity within a dominant plant outweighs plant species diversity in structuring an arthropod community. Ecology 94:1025–1035
Crous PW, Groenewald JZ, Pongpanich K, Himaman W, Arzanlou M, Wingfield MJ (2004) Cryptic speciation and host specificity among Mycosphaerella spp. occurring on Australian Acacia species grown as exotics in the tropics. Stud Mycol 50:457–469
Cruikshank MG, Jaquish B (2014) Resistance and tolerance in juvenile interior Douglas-fir trees Pseudotsuga menziesii var. glauca artificially inoculated with Armillaria ostoyae. For Pathol 44:362–371
Davelos AL, Jarosz AM (2004) Demography of American chestnut populations: effects of a pathogen and a hyperparasite. J Ecol 92:675–685
Dawkins R (1999) The extended phenotype. Oxford University Press, Oxford
Dawson JC, Goldringer I (2011) Breeding for genetically diverse populations: variety mixtures and evolutionary populations. In: Lammerts van Bueren ET, Myers JR (eds) Organic crop breeding. Wiley-Blackwell, Hoboken, pp 77–98
de Beer ZW, Duong TA, Barnes I, Wingfield BD, Wingfield MJ (2014) Redefining Ceratocystis and allied genera. Stud Mycol 79:187–219
de Vienne DM, Hood ME, Giraud T (2009) Phylogenetic determinants of potential host shifts in fungal pathogens. J Evol Biol 22:2532–2541
de Wet J, Burgess T, Slippers B, Preisig O, Wingfield BD, Wingfield MJ (2003) Multiple gene genealogies and microsatellite markers reflect relationships between morphotypes of Sphaeropsis sapinea and distinguish a new species of Diplodia. Mycol Res 107:557–566
Dehnen-Schmutz K, Holdenrieder O, Jeger MJ, Pautasso M (2010) Structural change in the international horticultural industry: some implications for plant health. Sci Hortic 125:1–15
Delatour C (1983) Les dépérissements de Chênes en Europe. Rev For France 35:262–282
Delatour C, Pinon J, Morelet M (1985) Histoire et avenir de la pathologie forestière en France. Rev For France 37:65–82
Delmotte F, Mestre P, Schneider C, Kassemeyer HH, Kozma P, Richart-Cervera S, Rouxel M, Delière L (2014) Rapid and multiregional adaptation to host partial resistance in a plant pathogenic oomycete: evidence from European populations of Plasmopara viticola, the causal agent of grapevine downy mildew. Infect Genet Evol 27:500–508
Denison RF, Kiers ET, West SA (2003) Darwinian agriculture: when can humans find solutions beyond the reach of natural selection? Q Rev Biol 78:145–168
Denman S, Mralejo E, Kirk S, et al (2008) Sporulation of Phytophthora ramorum and P. kernoviae on asymptomatic foliage and fruit. Proc Sudd Oak Death Third Sci Symp General Technical Report PSW-GTR-214: 201–207
Desprez-Loustau ML, Robin C, Buee M, Courtecuisse R, Garbaye J, Suffert F, Sache I, Rizzo DM (2007) The fungal dimension of biological invasions. Trends Ecol Evol 22:472–480
Desprez-Loustau ML, Courtecuisse R, Robin C, Husson C, Moreau PA, Blancard D, Selosse MA, Lung-Escarmant B, Piou D, Sache I (2010) Species diversity and drivers of spread of alien fungi (sensu lato) in Europe with a particular focus on France. Biol Invasions 12:157–172
Desprez-Loustau M-L, Feau N, Mougou-Hamdane A, Dutech C (2011) Interspecific and intraspecific diversity in oak powdery mildews in Europe: coevolution history and adaptation to their hosts. Mycoscience 52:163–175
Dhillon B, Feau N, Aerts AL, Beauseigle S, Bernier L, Copeland A, Foster A, Gill N, Henrissat B, Herath P, LaButti KM, Levasseur A, Lindquist EA, Majoor E, Ohm RA, Pangilinan JL, Pribowo A, Saddler JN, Sakalidis ML, de Vries RP, Grigoriev IV, Goodwin SB, Tanguay P, Hamelin RC (2015) Horizontal gene transfer and gene dosage drives adaptation to wood colonization in a tree pathogen. Proc Natl Acad Sci U S A 112:3451–3456
Di Bella JM, Bao Y, Gloor GB, Burton JP, Reid G (2013) High throughput sequencing methods and analysis for microbiome research. J Microbiol Methods 95:401–414
Dickmann DI, Gold MA, Flore JA (1994) The ideotype concept and the genetic improvement of tree crops. Plant Breed Rev 12: 163–193
Dlugosch K, Parker I (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Donald CM (1968) The breeding of crop ideotype. Euphytica 17:385–403
Dong S, Stam R, Cano LM, Song J, Sklenar J, Yoshida K, Bozkurt TO, Oliva R, Liu Z, Tian M, Win J, Banfield MJ, Jones AM, van der Hoorn RA, Kamoun S (2014) Effector specialization in a lineage of the Irish potato famine pathogen. Science 343:552–555
Donnelly A, Caffarra A, Kelleher CT, O'neill BF, Diskin E, Pletsers A, Proctor H, Stirnemann R, O'Halloran J, Peñuelas J, Hodkinson TR, Sparks TH (2012) Surviving in a warmer world: environmental and genetic responses. Clim Res 53:245–262
Dowkiw A, Jorge V, Villar M, Voisin E, Guérin V, Faivre-Rampant P, Bresson A, Bitton F, Duplessis S, Frey P, Petre B, Guinet C, Xhaard C, Fabre B, Halkett F, Plomion C, Lalanne C, Bastien C (2012) Breeding poplars with durable resistance to Melampsora larici-populina leaf rust: a multidisciplinary approach to understand and delay pathogen adaptation. In: Sniezko RA et al (eds) Proceedings of the fourth international workshop on the genetics of host-parasite interactions in forestry: disease and insect resistance in forest trees, Gen. Tech. Rep. PSW-GTR-240. Pacific Southwest Research Station, Forest Service, U.S. Department of Agriculture, Albany, pp 31–38
Dubouzet JG, Strabala TJ, Wagner A (2013) Potential transgenic routes to increase tree biomass. Plant Sci 212:72-101
Dutech C, Fabreguettes O, Capdevielle X, Robin C (2010) Multiple introductions of divergent genetic lineages in an invasive fungal pathogen, Cryphonectria parasitica, in France. Heredity 105:220–228
Dutech C, Barres B, Bridier J, Robin C, Milgroom MG, Ravigne V (2012) The chestnut blight fungus world tour: successive introduction events from diverse origins in an invasive plant fungal pathogen. Mol Ecol 21:3931–3946
Elegbede CF, Pierrat J-C, Aguayo J, Husson C, Halkett F, Marçais B (2010) A statistical model to detect asymptomatic infectious individuals with an application in the Phytophthora alni-induced alder decline. Phytopathology 100:1262–1269
Ellen ED, Rodenburg TB, Albers GA, Bolhuis JE, Camerlink I, Duijvesteijn N, Knol EF, Muir WM, Peeters K, Reimert I, Sell-Kubiak E, van Arendonk JA, Visscher J, Bijma P (2014) The prospects of selection for social genetic effects to improve welfare and productivity in livestock. Front Genet 5:377
Elton CS (1958) The ecology of invasions by animals and plants, University of Chicago Press edth edn. University of Chicago Press, Chicago
Ennos RA (2001) The introduction of lodgepole pine as a major forest crop in Sweden: implications for host–pathogen evolution. For Ecol Manag 141:85–96
Ennos RA (2015) Resilience of forests to pathogens: an evolutionary ecology perspective. Forestry 88:41–52
Eschen R, Rigaux L, Sukovata L, Vettraino AM, Marzano M, Gregoire JC (2015) Phytosanitary inspection of woody plants for planting at European Union entry points: a practical enquiry. Biol Invasions. doi:10.1007/s10530-015-0883-6
Essl F, Dullinger S, Rabitsch W, Hulme PE, Hülber K, Jarosík V, Kleinbauer I, Krausmann F, Kühn I, Nentwig W, Vilà M, Genovesi P, Gherardi F, Desprez-Loustau ML, Roques A, Pysek P (2011) Socioeconomic legacy yields an invasion debt. Proc Natl Acad Sci U S A 108:203–207
Fabre B, Ioos R, Piou D, Marçais B (2011) Is the emergence of Dothistroma needle blight of pine in France caused by the cryptic species Dothistroma pini? Phytopathology 102:47–54
Facon B, Genton B, Shykoff J, Jarne P, Estoup A, David P (2006) A general eco-evolutionary framework for understanding bioinvasions. Trends Ecol Evol 21:130–135
FAO (2010) Global forest resources assessment, FAO Forestry Paper 163, Rome
Faust K, Raes J (2012) Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550
Fauvergue X, Vercken E, Malausa T, Hufbauer RA (2012) The biology of small, introduced populations, with special reference to biological control. Evol Appl 5:424–443
Fears R, Aroemail EM, Pais MS, ter Meulen V (2014) How should we tackle the global risks to plant health? Trends Plant Sci 19:206–208
Feau N, Lauron-Moreau A, Piou D, Marçais B, Dutech C, Desprez-Loustau ML (2012) Niche partitioning of genetic lineages involved in the oak powdery mildew complex. Fungal Ecol 5:154–162
Feau N, Dutech C, Brusini J, Rigling D, Robin C (2014) Multiple introductions and recombination in Cryphonectria hypovirus 1: perspective for a sustainable biological control of chestnut blight. Evol Appl 7:580–596
Fine PVA, Miller ZJ, Mesones I, Irazuzta S, Appel HM, Stevens MHH, Saaksjarvi I, Schultz LC, Coley PD (2006) The growth defense trade-off and habitat specialization by plants in Amazonian forests. Ecology 87:S150–S162
Fitt BDL, Huang YJ, van den Bosch F, West JS (2006) Coexistence of related pathogen species on arable crops in space and time. Annu Rev Phytopathol 44:163–182
Fortuna MA, Zaman L, Wagner AP, Ofria C (2013) Evolving digital ecological networks. PLoS Comput Biol 9, e1002928. doi:10.1371/journal.pcbi.1002928
Franceschini S, Webber JF, Sancisi-Frey S, Brasier CM (2014) Gene × environment tests discriminate the new EU2 evolutionary lineage of Phytophthora ramorum and indicate that it is adaptively different. For Pathol 44:219–232
Frank SA (1993) Evolution of host–parasite diversity. Evolution 47:1721–1732
Frank S (1996) Models of parasite virulence. Q Rev Biol 71:37–78
Frankham R (2005) Invasion biology—resolving the genetic paradox in invasive species. Heredity 94:385
Franklin O, Palmroth S, Nasholm T (2014) How eco-evolutionary principles can guide tree breeding and tree biotechnology for enhanced productivity. Tree Physiol 34:1149–1166
Gandon S (2002) Local adaptation and the geometry of host–parasite coevolution. Ecol Lett 5:246–256
Gandon S, Michalakis Y (2002) Local adaptation, evolutionary potential and host-parasite coevolution: interactions between migration, mutation, population size and generation time. J Evol Biol 15:451–462
Garbelotto M (2008) Molecular analysis to study invasions by forest pathogens: examples from Mediterranean ecosystems. Phytopathol Mediterr 47:183–203
Garbelotto M, Linzer R, Nicolotti G, Gonthier P (2010) Comparing the influences of ecological and evolutionary factors on the successful invasion of a fungal forest pathogen. Biol Invasions 12:943–957
García-Guzmán G, Heil M (2014) Life histories of hosts and pathogens predict patterns in tropical fungal plant diseases. New Phytol 201:1106–1120
Ghabrial SA, Suzuki N (2009) Viruses of plant pathogenic fungi. Annu Rev Phytopathol 47:353–384
Gilbert G (2002) Evolutionary ecology of plant diseases in natural ecosystems. Annu Rev Phytopathol 40:13–43
Gilbert GS, Webb CO (2007) Phylogenetic signal in plant pathogen–host range. Proc Natl Acad Sci U S A 104:4979–4983
Gilbert GS, Magarey R, Suiter K, Webb CO (2012) Evolutionary tools for phytosanitary risk analysis: phylogenetic signal as a predictor of host range of plant pests and pathogens. Evol Appl 5:869–878
Gilligan CA (2008) Sustainable agriculture and plant diseases: an epidemiological perspective. Philos Trans R Soc Lond B Biol Sci 363:741–759
Giraud T, Enjalbert J, Fournier E, Delmotte F, Dutech C (2008a) Population genetics of fungal diseases of plants. Parasite 15:449–454
Giraud T, Refrégier G, Le Gac M, De Vienne DM, Hood ME (2008b) Speciation in fungi. Fungal Genet Biol 45:791–802
Giraud T, Gladieux P, Gavrilets S (2010) Linking the emergence of fungal plant diseases with ecological speciation. Trends Ecol Evol 25:387–395
Gladieux P, Feurtey A, Hood ME, Snirc A, Clavel J, Dutech C, Roy M, Giraud T (2015) The population biology of fungal invasions. Mol Ecol. doi:10.1111/mec.13028
Goss EM, Larsen M, Chastagner GA, Givens DR, Grünwald NJ (2009) Population genetic analysis infers migration pathways of Phytophthora ramorum in US nurseries. PLoS Pathog 5, e1000583
Goss EM, Tabima JF, Cooke DEL, Restrepo S, Fry WE, Forbes GA, Fieland VJ, Cardenas M, Grünwald NJ (2014) The Irish potato famine pathogen Phytophthora infestans originated in central Mexico rather than the Andes. Proc Natl Acad Sci U S A 111:8791–8796
Graça RN, Ross-Davis AL, Klopfenstein NB, Kim M-S, Peever TL, Cannon PG, Aun CP, Mizubuti ESG, Alfenas AC (2013) Rust disease of eucalypts, caused by Puccinia psidii, did not originate via host jump from guava in Brazil. Mol Ecol 22:6033–6047
Grente J (1981) Les variants hypovirulents de l’Endothia parasitica et la lutte biologique contre le chancre du châtaignier. Université de Bretagne Occidentale, Brest, France, Dissertation
Gross A, Hosoya T, Queloz V (2014) Population structure of the invasive forest pathogen Hymenoscyphus pseudoalbidus. Mol Ecol 23:2943–2960
Grünwald NJ, Goss EM (2011) Evolution and population genetics of exotic and re-emerging pathogens: novel tools and approaches. Annu Rev Phytopathol 49:249–267
Hacquard S, Schadt CW (2015) Towards a holistic understanding of the beneficial interactions across the Populus microbiome. New Phytol 205:1425–1430
Hale IL, Broders K, Iriarte G (2014) A Vavilovian approach to discovering crop-associated microbes with potential to enhance plant immunity. Front Plant Sci 5:492
Hamelin F, Castel M, Poggi S, Andrivon D, Mailleret L (2011) Seasonality and the evolutionary divergence of plant parasites. Ecology 92:2159–2166
Hansen EM (2008) Alien forest pathogens: Phytophthora species are changing world forests. Boreal Environ Res 13:33–41
Hansen E, Goheen E (2000) Phellinus weirii and other native root pathogens as determinants of forest structure and process in western North America. Annu Rev Phytopathol 38:515–539
Hansen EM, Parke JL, Sutton W (2005) Susceptibility of Oregon forest trees and shrubs to Phytophthora ramorum: a comparison of artificial inoculation and natural infection. Plant Dis 89:63–70
Hansen EM, Reeser PW, Sutton W (2011) Phytophthora beyond agriculture. Annu Rev Phytopathol 50:359–378
Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH (2012) Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10:497–506
Hantsch L, Braun U, Scherer-Lorenzen M, Bruelheide H (2013) Species richness and species identity effects on occurrence of foliar fungal pathogens in a tree diversity experiment. Ecosphere 4:81
Hantsch L, Braun U, Haase J, Purschke O, Scherer-Lorenzen M, Bruelheide H (2014) No plant functional diversity effects on foliar fungal pathogens in experimental tree communities. Fungal Divers 66:139–151
Harfouche A, Meilan R, Kirst M, Morgante M, Boerjan W, Sabatti M, ScarasciaMugnozza G (2012) Accelerating the domestication of forest trees in a changing world. Trends Plant Sci 17:64–72
Harwood CE, Nambiar EKS (2014) Productivity of acacia and eucalypt plantations in Southeast Asia. 2. Trends and variations. Int For Rev 16:249–260
Hayden K, Nettel A, Dodd RS, Garbelotto M (2011) Will all the trees fall? Variable resistance to an introduced forest disease in a highly susceptible host. For Ecol Manag 261:1781–1791
Hayden K, Hardy G, Garbelotto M (2013) Oomycete diseases. In: Gonthier P, Nicolotti G (eds) Infectious forest diseases. CABI, Boston, pp 519–546
Hendry AP, Kinnison MT, Heino M, Day T, Smith TB, Fitt G, Bergstrom CT, Oakeshott J, Jørgensen PS, Zalucki MP, Gilchrist G, Southerton S, Sih A, Strauss S, Denison RF, Carroll SP (2011) Evolutionary principles and their practical application. Evol Appl 4:159–183
Hepting GH (1963) Climate and forest diseases. Annu Rev Phytopathol 1:31–50
Horton MW, Bodenhausen N, Beilsmith K, Meng D, Muegge BD, Subramanian S, Vetter MM, Vilhjálmsson BJ, Nordborg M, Gordon JI, Bergelson J (2014) Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat Commun 10:5320
Hüberli D, Hardy GESJ, White D, Williams N, Burgess TI (2013) Fishing for Phytophthora from Western Australia’s waterways: a distribution and diversity survey. Australas Plant Pathol 42:251–260
Hudson PJ, Dobson AP, Lafferty KD (2006) Is a healthy ecosystem one that is rich in parasites? Trends Ecol Evol 21:381–385
Hufbauer RA, Roderick GK (2005) Microevolution in biological control: mechanisms, patterns, and processes. Biol Control 35:227–239
Husson C, Scala B, Caël O, Frey P, Feau N, Ioos R, Marçais B (2011) Chalara fraxinea is an invasive pathogen in France. Eur J Plant Pathol 130:311–324
Husson C, Aguayo J, Revellin C, Frey P, Ioos R, Marçais B (2015) Evidence for homoploid speciation in Phytophthora alni supports taxonomic reclassification in this species complex. Fungal Genet Biol 77:12–21
Hyde KD (2001) Where are the missing fungi? Does Hong Kong have any answers? Mycol Res 105:1514–1518
Ioos R, Andrieux A, Marcais B, Frey P (2006) Genetic characterization of the natural hybrid species Phytophthora alni as inferred from nuclear and mitochondrial DNA analyses. Fungal Genet Biol 43:511–529
Jarosz AM, Davelos AL (1995) Effects of disease in wild plant-populations and the evolution of pathogen aggressiveness. New Phytol 129:371–387
Jeger MJ, Pautasso M (2008) Plant disease and global change—the importance of long-term data sets. New Phytol 177:8–11
Johnson R (1984) A critical analysis of durable resistance. Annu Rev Phytopathol 22:309–330
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329
Jousimo J, Tack AJM, Ovaskainen O, Mononen T, Susi H, Tollenaere C, Laine A-L (2014) Ecological and evolutionary effects of fragmentation on infectious disease dynamics. Science 344:1289–1293
Jumpponen A, Jones KL (2009) Massively parallel 454 sequencing indicates hyperdiverse fungal communities in temperate Quercus macrocarpa phyllosphere. New Phytol 184:438–448
Jumpponen A, Jones KL (2010) Seasonally dynamic fungal communities in the Quercus macrocarpa phyllosphere differ between urban and nonurban environments. New Phytol 186:496–513
Kamgan NG, Jacobs K, de Beer ZW, Wingfield MJ, Roux J (2008) Ceratocystis and Ophiostoma species, including three new taxa, associated with wounds on native South African trees. Fungal Divers 29:37–59
Keesing F, Holt RD, Ostfeld RS (2006) Effects of species diversity on disease risk. Ecol Lett 9:485–498
Keller SR, Taylor DR (2008) History, chance, and adaptation during biological invasion: separating stochastic phenotypic evolution from response to selection. Ecol Lett 11:852–866
Kembel SW, Mueller RC (2014) Plant traits and taxonomy drive host associations in tropical phyllosphere fungal communities. Botany 92:303–311
Kembel SW, O’Connor TK, Arnold HK, Hubbell SP, Wright SJ, Green JL (2014) Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc Natl Acad Sci 111:13715–13720
Kemen E (2014) Microbe-microbe interactions determine oomycete and fungal host colonization. Curr Opin Plant Biol 20:75–81
Kempel A, Schadler M, Chrobock T, Fischer M, van Kleunen M (2011) Tradeoffs associated with constitutive and induced plant resistance against herbivory. Proc Natl Acad Sci U S A 108:5685–5689
Kim MS, Klopfenstein NB, Ota Y, Lee SK, Woo KS, Kaneko S (2010) White pine blister rust in Korea, Japan and other Asian regions: comparisons and implications for North America. For Pathol 40:382–401
King KC, Lively CM (2012) Does genetic diversity limit disease spread in natural host populations? Heredity 109:199–203
Kinkel LL, Bakker MG, Schlatter DC (2011) A coevolutionaryframework for managing disease-suppressive soils. Annu Rev Phytopathol 49:47–67
Kinloch B, Sniezko R, Dupper G (2004) Virulence gene distribution and dynamics of the white pine blister rust pathogen in western North America. Phytopathology 94:751–758
Kinloch B, Davis D, Burton D (2008) Resistance and virulence interactions between two white pine species and blister rust in a 30-year field trial. Tree Genet Genomes 4:65–74
Kiss L (2003) A review of fungal antagonists of powdery mildews and their potential as biocontrol agents. Pest Manag Sci 59:475–483
Kliebenstein DJ (2014) Orchestration of plant defense systems: genes to populations Daniel J. Trends Plant Sci 19:250–255
Knief C, Ramette A, Frances L, Alonso-Blanco C, Vorholt JA (2010) Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. ISME J 4:719–728
Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, von Mering C, Vorholt JA (2012) Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J 6: 1378–1390
Knops JMH, Tilman D, Haddad NM, Naeem S, Mitchell CE, Haarstad J, Ritchie ME, Howe KM, Reich PB, Siemann E, Groth J (1999) Effects of plant species richness on invasion dynamics, disease outbreaks, insect abundances and diversity. Ecol Lett 2:286–293
Koch H, Frickel J, Valiadi M, Becks L (2014) Why rapid, adaptive evolution matters for community dynamics. Front Ecol Evol 2:17
Kolbe JJ, Glor RE, Rodríguez Schettino L, Lara AC, Larson A, Losos JB (2004) Genetic variation increases during biological invasion by a Cuban lizard. Nature 431:177–181
Koskella B (2013) Phage-mediated selection on microbiota of a long-lived host. Curr Biol 23:1256–1260
Koskella B (2014) Bacteria-phage interactions across time and space: merging local adaptation and time-shift experiments to understand phage evolution. Am Nat 184:S9–S21
Koskella B, Lin DM, Buckling A, Thompson JN (2012) The costs of evolving resistance in heterogeneous parasite environments. Proc R Soc B 279:1896–1903
Kroon LPNM, Brouwer H, de Cock AWAM, Govers F (2011) The genus Phytophthora Anno 2012. Phytopathology 102:348–364
Kupferschmidt K (2012) Attack of the clones. Science 337:636–638
Kutschera U, Hossfeld U (2012) Physiological phytopathology: origin and evolution of a scientific discipline. J Appl Bot Food Qual 85:1–5
Kvas M, Marasas WFO, Wingfield BD, Wingfield MJ, Steenkamp ET (2009) Diversity and evolution of Fusarium species in the Gibberella fujikuroi complex. Fungal Divers 34:1–21
Lafferty KD, Dobson AP, Kuris AM (2006) Parasites dominate food web links. Proc Natl Acad Sci U S A 103:11211–11216
Lambrechts L, Fellous S, Koella JC (2006) Coevolutionary interactions between host and parasite genotypes. Trends Parasitol 22:12–16
Lavergne S, Molofsky J (2007) Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc Natl Acad Sci U S A 104:3883–3888
Le Gac M, Giraud T (2008) Existence of a pattern of reproductive character displacement in Homobasidiomycota but not in Ascomycota. J Evol Biol 21:761–772
Lebeis SL (2014) The potential for give and take in plant–microbiome relationships Front. Plant Sci 5:287
Lee CE (2002) Evolutionary genetics of invasive species. Trends Ecol Evol 17:386–391
Lefevre F, Boivin T, Bontemps A, Courbet F, Davi H, Durand-Gillmann M, Fady B, Gauzere J, Gidoin C, Karam MJ, Lalagüe H, Oddou-Muratorio S, Pichot C (2014) Considering evolutionary processes in adaptive forestry. Ann For Sci 71:723–739
Lieberei R (2007) South American leaf blight of the rubber tree (Hevea spp.): New steps in plant domestication using physiological features and molecular markers. Ann Bot 100:1125–1142
Liebhold AM, Wingfield MJ (2014) Globalization and its implications to forest health. In: Nikolakis W, Innes J (eds) Forests and globalization: challenges and opportunities for sustainable development. Routledge, pp 36–47
Liebhold AM, Brockerhoff EG, Garrett LJ, Parke JL, Britton KO (2012) Live plant imports: the major pathway for forest insect and pathogen invasions of the U.S. Front Ecol Environ 10:135–143
Lind EM, Borer E, Seabloom E, Adler P, Bakker JD, Blumenthal DM, Crawley M, Davies K, Firn J, Gruner DS, Harpole WS, Hautier Y, Hillebrand H, Knops J, Melbourne B, Mortensen B, Risch AC, Schuetz M, Stevens C, Wragg PD (2013) Life-history constraints in grassland plant species: a growth-defence trade-off is the norm. Ecol Lett 16:513–521
Linzer R, Rizzo D, Cacciola S, Garbelotto M (2009) AFLPs detect low genetic diversity for Phytophthora nemorosa and P. pseudosyringae in the US and Europe. Mycol Res 113:298–307
Little TJ, Shuker DM, Colegrave N, Day T, Graham AL (2010) The coevolution of virulence: tolerance in perspective. PLoS Pathog 6, e1001006. doi:10.1371/journal.ppat.1001006
Lively CM, de Roode JC, Duffy MA, Graham AL, Koskella B (2014) Interesting open questions in disease ecology and evolution. Am Nat 184:S1–S8
Loehle C, Namkoong G (1987) Constraints on tree breeding: growth tradeoffs, growth strategies, and defensive investments. For Sci 33:1089–1097
Loo JA (2009) Ecological impacts of non-indigenous invasive fungi as forest pathogens. Biol Invasions 11:81–96
Manion PD (1981) Tree disease concepts. Prentice-Hall, Englewood Cliffs, 399 pp
Manion PD (2003) Evolution of concepts in forest pathology. Phytopathology 93:1052–1055
Martin TA, Johnsen KH, White TL (2001) Ideotype Development in Southern Pines: Rationale and Strategies for Overcoming Scale-Related Obstacles. For Sci 47: 21-28
Martinez-Alvarez P, Vainio EJ, Botella L, Hantula J, Diez JJ (2014) Three mitovirus strains infecting a single isolate of Fusarium circinatum are the first putative members of the family Narnaviridae detected in a fungus of the genus Fusarium. Arch Virol 159:2153–2155
Matos A, Kerkhof L, Garland JL (2005) Effects of microbial community diversity on the survival of Pseudomonas aeruginosa in the wheat rhizosphere. Microb Ecol 49:257–264
May RM, Anderson RM (1983) Epidemiology and genetics in the coevolution of parasites and hosts. Proc R Soc B 219:281–313
McCracken AR, Walsh LRE, Moore JP, Lynch M, Cowan P, Dawson MD, Watson S (2011) Yield of willow (Salix spp.) grown in short rotation coppice mixtures in long-term trials. Ann Appl Biol 159:229–243
McDonald BA, Linde C (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 40:349–379
McKown AD, Guy RD, Quamme L, Klápště J, La Mantia J, Constabel CP, El-Kassaby YA, Hamelin RC, Zifkin M, Azam MS (2014) Association genetics, geography and ecophysiology link stomatal patterning in Populus trichocarpa with carbon gain and disease resistance trade-offs. Mol Ecol 23: 5771-90
Merrill W, Shigo AL (1979) An expanded concept of tree decay. Phytopathology 69:1158–1160
Michelmore RW, Christopoulou M, Caldwell KS (2013) Impacts of resistance gene genetics, function, and evolution on a durable future. Annu Rev Phytopathol 51:291–319
Milgroom MG, Cortesi P (2004) Biological control of chestnut blight with hypovirulence: a critical analysis. Annu Rev Phytopathol 42:311–338
Millennium Ecosystem Assessment (2005) Ecosystems and human well-being: synthesis. Island Press, Washington, DC
Miller MR, White A, Boots M (2006) The evolution of parasites in response to tolerance in their hosts: the good, the bad, and apparent commensalism. Evol Int J Org Evol 60:945–956
Milus EA, Kristensen K, Hovmoller MS (2009) Evidence for increased aggressiveness in a recent widespread strain of Puccinia striiformis f. sp. tritici causing stripe rust of wheat. Phytopathology 99:89–94
Mitchell CE, Power AG (2003) Release of invasive plants from fungal and viral pathogens. Nature 421:625–627
Mohan V, Manokaran P (2013) Assessment of disease problems in different clonal plantations of Eucalyptus spp. in South India. J Acad Ind Res 1:514–524
Morozov A, Robin C, Franc A (2007) A simple model for the dynamics of a host–pathogen–hyperparasite interaction. J Theor Biol 249:246–253
Mosquera J, Adler FR (1998) Evolution of virulence: a unified framework for coinfection and superinfection. J Theor Biol 195:293–313
Mougou A, Dutech C, Desprez-Loustau M-L (2008) New insights into the identity and origin of the causal agent of oak powdery mildew in Europe. For Pathol 38:275–287
Mougou-Hamdane A, Giresse X, Dutech C, Desprez-Loustau ML (2010) Spatial distribution of lineages of oak powdery mildew fungi in France, using quick molecular detection methods. Ann For Sci 67:212
Mukherjee PK, Horwitz BA, Herrera-Estrella A, Schmoll M, Kenerley CM (2013) Trichoderma research in the genome era. Annu Rev Phytopathol 51:105–129
Neale D, Ingvarsson P (2008) Population, quantitative and comparative genomics of adaptation in forest trees. Curr Opin Plant Biol 11:149–155
Newcombe G, Dugan FM (2010) Fungal pathogens of plants in the Homogocene. In: Gherbawy Y, Voigt K (eds) Molecular identification of fungi. Springer-Verlag, Berlin, pp 3–35
Newcombe G, Stirling B, McDonald S, Bradshaw HD Jr (2000) Melampsora × columbiana, a natural hybrid of M. medusae and M occidentalis. Mycol Res 104:261–274
Newton AC, Gravouil C, Fountaine JM (2010) Managing the ecology of foliar pathogens: ecological tolerance in crops. Ann Appl Biol 157:343–359
Ney B, Bancal MO, Bancal P, Bingham IJ, Foulkes J, Gouache D, Paveley N, Smith J (2013) Crop architecture and crop tolerance to fungal diseases and insect herbivory. Mechanisms to limit crop losses. Eur J Plant Pathol 135:561–580
Oh E, Gryzenhout M, Wingfield BD, Wingfield MJ, Burgess TI (2013) Surveys of soil and water reveal a goldmine of Phytophthora diversity in South African natural ecosystems. IMA Fungus 4:123–131
Oliva J, Camarero JJ, Stenlid J (2012) Understanding the role of sapwood loss and reaction zone formation on radial growth of Norway spruce (Picea abies) trees decayed by Heterobasidion annosum s.l. For Ecol Manag 274:201–209
Oliva J, Stenlid J, Martinez-Vilalta J (2014) The effect of fungal pathogens on the water and carbon economy of trees: implications for drought-induced mortality. New Phytol 203:1028–1035
O'Malley M (2008) ‘Everything is everywhere, but the environment selects’: ubiquitous distribution and ecological determinism in microbial biogeography. Stud Hist Phil Biol Biomed Sci 39:314–325
Ostry ME, Laflamme G (2009) Fungi and diseases—natural components of healthy forests. Botany 87:22–25
Palumbi SR (2001) Humans as the world’s greatest evolutionary force. Science 293:1786–1790
Papaïx J, Burdon JJ, Zhan J, Thrall PH (2015) Crop pathogen emergence and evolution in agro-ecological landscapes. Evol Appl. doi:10.1111/eva.12251
Paquette A, Messier C (2009) The role of plantations in managing the world’s forests in the Anthropocene. Front Ecol Environ 8:27–34
Parke JL, Grünwald NJ (2012) A systems approach for management of pests and pathogens of nursery crops. Plant Dis 96:1236–1244
Parke JL, Knaus BJ, Fieland VJ, Lewis C, Grünwald NJ (2014) Phytophthora community structure analyses in Oregon nurseries inform systems approaches to disease management. Phytopathology 104:1052–1062
Parker IM, Gilbert GS (2004) The evolutionary ecology of novel plant–pathogen interactions. Annu Rev Ecol Evol Systematics 35:675–700
Parker JD, Burkepile DE, Hay ME (2006) Opposing effects of native and exotic herbivores on plant invasions. Science 311:1459–1461
Partida-Martínez LP, Heil M (2011) The microbe-free plant: fact or artifact? Front Plant Sci 2:100
Pautasso M, Holdenrieder O, Stenlid, J (2005) Susceptibility to fungal pathogens of forests differing in tree diversity. In: Scherer-Lorenzen M, Körner C, Schulze E-D (Eds.) Forest diversity and function: temperate and boreal systems. Springer, pp 263–289
Pautasso M, Schlegel M, Holdenrieder O (2014) Forest health in a changing world. Microb Ecol. doi:10.1007/s00248-014-0545-8
Pearson MN, Beever RE, Boine B, Arthur K (2009) Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol 10:115–128
Peay K, Baraloto C, Fine PVA (2013) Strong coupling of plant and fungal structure across Amazonian rainforests. ISME J 7:1851–1861
Peiffer J, Spor A , Jin Z, Koren O, Tringe SG, Dangl JL, Buckler ES, Ley RE (2013) Diversity and heritability of the Maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA 5: 570-573
Peñuelas J, Rico L, Ogaya R, Jump AS, Terradas J (2012) Summer season and long-term drought increase the richness of bacteria and fungi in the foliar phyllosphere of Quercus ilex in a mixed Mediterranean forest. Plant Biol (Stuttg) 14:565–575
Pérez G, Slippers B, Wingfield MJ, Wingfield BD, Carnegie AJ, Burgess TI (2012) Cryptic species, native populations and biological invasions by a eucalypt forest pathogen. Mol Ecol 21:4452–4471
Peterson PD, Griffith CS (1999) Hermann von Schrenk and the beginning of forest pathology in the US. Forest History Today, 29–34
Pinon J, Frey P (2005) Interactions between poplar clones and Melampsora populations and their implications for breeding for durable resistance. In: Pei MH, McCracken AR (eds) Rust diseases of willow and poplar. CAB International, Wallingford, pp 139–154
Power AG, Mitchell CE (2004) Pathogen spillover in disease epidemics. Am Nat 164:S79–S89
Prospero S, Conedera M, Heiniger U, Rigling D (2006) Saprophytic activity and sporulation of Cryphonectria parasitica on dead chestnut wood in forests with naturally established hypovirulence. Phytopathology 96:1337–1344
Pyšek P, Jarošíkb V, Hulme PE, Kühn I, Wilda J, Arianoutsou M, Bacher S, Chiron F, Didžiulisi D, Essl F, Genovesi P, Gherardi F, Hejda M, Kark S, Lambdon PW, Desprez-Loustau M-L, Nentwig W, Pergl J, Poboljša K, Rabitsch W, Roques A, Roy DB, Shirley S, Solarz W, Vilà M, Winter M (2010) Disentangling the role of environmental and human pressures on biological invasions. Proc Natl Acad Sci U S A 107:12157–12162
Queloz V, Grünig CR, Berndt R, Kowalski T, Sieber TN, Holdenrieder O (2011) Cryptic speciation in Hymenoscyphus albidus. For Pathol 41:133–142
Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N (2010) The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ Microbiol 12:2885–2893
Refregier G, Le Gac M, Jabbour F, Widmer A, Shykoff JA, Yockteng R, Giraud T (2008) Co-phylogeny of the anther smut fungi and their caryophyllaceous hosts: prevalence of host shifts and importance of delimiting parasite species for inferring co-speciation. BMC Evol Biol 8:10
Restif O, Koella JC (2004) Concurrent evolution of resistance and tolerance to pathogens. Am Nat 164:E90–E102
REX Consortium (2013) Heterogeneity of selection and the evolution of resistance. Trends Ecol Evol 28:110–118
Rico L, Ogaya R, Terradas J, Peñuelas J (2014) Community structures of N2 -fixing bacteria associated with the phyllosphere of a Holm oak forest and their response to drought. Plant Biol 16:586–593
Ridout N, Newcombe G (2015) The frequency of modification of Dothistroma pine needle blight severity by fungi within the native range. For Ecol Manag 337:153–160
Rius M, Darling JA (2014) How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol Evol 29:233–242
Robin C, Lanz S, Soutrenon A, Rigling D (2010) Dominance of natural over released biological control agents of the chestnut blight fungus Cryphonectria parasitica in south-eastern France is associated with fitness-related traits. Biol Control 53:55–61
Roderick GK, Navajas M (2003) Genes in new environments: genetics and evolution in biological control. Nat Genet 4:889–899
Roderick GK, Hufbauer R, Navajas M (2012) Evolution and biological control. Evol Appl 5:419–423
Roossinck MJ, Saha P, Wiley GB, Quan J, White JD, Lai H, Chavarria F, Shen GA, Roe BA (2010) Ecogenomics: using massively parallel pyrosequencing to understand virus ecology. Mol Ecol 19:81–88
Roper M, Ellison C, Taylor JW, Glass NL (2011) Nuclear and genome dynamics in multinucleate ascomycete fungi. Curr Biol 21:R786–R793
Roy BA (2001) Patterns of association between crucifers and their flower-mimic pathogens: host-jumps are more common than coevolution or cospeciation. Evolution 55:41–53
Roy BA, Kirchner JW (2000) Evolutionary dynamics of pathogen resistance and tolerance. Evolution 54:51–63
Roy BA, Alexander HM, Davidson J, Campbell FT, Burdon JJ, Sniezko R, Brasier C (2014) Increasing forest loss worldwide from invasive pests requires new trade regulations. Front Ecol Environ 12:457–465
Sakalidis ML, Slippers B, Wingfield BD, Hardy GESJ, Burgess TI (2013) The challenge of understanding the origin, pathways and extent of fungal invasions: global populations of the Neofusicoccum parvum–N ribis species complex. Divers Distrib 19:873–883
Santini A, Ghelardini L, De Pace C, Desprez-Loustau ML, Capretti P, Chandelier A, Cech T, Chira D, Diamandis S, Gaitniekis T, Hantula J, Holdenrieder O, Jankovsky L, Jung T, Jurc D, Kirisits T, Kunca A, Lygis V, Malecka M, Marcais B, Schmitz S, Schumacher J, Solheim H, Solla A, Szabò I, Tsopelas P, Vannini A, Vettraino AM, Webber J, Woodward S, Stenlid J (2013) Biogeographical patterns and determinants of invasion by forest pathogens in Europe. New Phytol 197:238–250
Schafer JF (1971) Tolerance to plant disease. Ann Rev Phytopathol 9:235–252
Schardl CL, Craven KD (2003) Interspecific hybridization in plant-associated fungi and oomycetes: a review. Mol Ecol 12:2861–2873
Schmit J, Mueller G (2007) An estimate of the lower limit of global fungal diversity. Biodivers Conserv 16:99–111
Schoebel CN, Zoller S, Rigling D (2014) Detection and genetic characterisation of a novel mycovirus in Hymenoscyphus fraxineus, the causal agent of ash dieback. Infect Genet Evol 28:78–86
Schoettle AW, Sniezko RA (2007) Proactive intervention to sustain high-elevation pine ecosystems threatened by white pine blister rust. J For Res 12:327–336
Schulenburg H, Kurtz J, Moret Y, Siva-Jothy MT (2009) Introduction. Ecological immunology. Phil Trans R Soc B 364:3–14
Schulze-Lefert P, Panstruga R (2011) A molecular concept connecting nonhost-resistance and pathogen speciation. Trends Plant Sci 16:117–125
Schweitzer JA, Bailey JK, Fischer DG, LeRoy CJ, Lonsdorf EV, Whitham TG, Hart SC (2008) Plant–soil–microorganism interactions: heritable relationship betwen plant genotype and associated soil microorganisms. Ecology 89:773–781
Selakovic S, de Ruiter PC, Heesterbeek H (2014) Infectious disease agents mediate interaction in food webs and ecosystems. Proc R Soc B 281:20132709. doi:10.1098/rspb.2013.2709
Shaw MW, Osborne TM (2011) Geographic distribution of plant pathogens in response to climate change. Plant Pathol 60:31–43
Shaw MW, Bearchell SJ, Fitt BDL, Fraaije BA (2008) Long-term relationships between environment and abundance in wheat of Phaeosphaeria nodorum and Mycosphaerella graminicola. New Phytol 177:229–238
Simms E (2000) Defining tolerance as a norm of reaction. Evol Ecol 14:563–570
Simms E, Triplett J (1994) Costs and benefits of plant responses to disease - resistance and tolerance. Evolution 48:1973–1985
Siozios S, Tosi L, Ferrarini A, Ferrari A, Tononi P, Bellin D, Maurhofer M, Gessler C, Delledonne M, Pertot I (2015) Transcriptional reprogramming of the mycoparasitic fungus Ampelomyces quisqualis during the powdery mildew host-induced germination. Phytopathology 105:199–209
Springer JC, Baines ALD, Fulbright DW, Chansler MT, Jarosz AM (2013) Hyperparasites influence population structure of the chestnut blight pathogen, Cryphonectria parasitica. Phytopathology 103:1280–1286
Stenlid J, Oliva J, Boberg JB, Hopkins AJM (2011) Emerging diseases in European forest ecosystems and responses in society. Forests 2:486–504
Stukenbrock EH (2013) Evolution, selection and isolation: a genomic view of speciation in fungal plant pathogens. New Phytol 199:895–907
Stukenbrock EH, Bataillon T (2012) A population genomics perspective on the emergence and adaptation of new plant pathogens in agro-ecosystems. PLoS Pathog 8, e1002893
Stukenbrock EH, McDonald BA (2008) The origins of plant pathogens in agro-ecosystems. Annu Rev Phytopathol 46:75–100
Sturrock RN, Frankel SJ, Brown AV, Hennon PE, Kliejunas JT, Lewis KJ, Woods AJ (2011) Climate change and forest diseases. Plant Pathol 60:133–149
Su-See L (1999) Forest health in plantation forests in South-East Asia. Australas Plant Pathol 28:283–291
Susi H, Barres B, Pedro F, Vale PF, Laine AL (2014) Co-infection alters population dynamics of infectious disease. Nat Commun. doi:10.1038/ncomms6975
Swett CL, Gordon TR (2012) First report of grass species (Poaceae) as naturally occurring hosts of the pine pathogen Gibberella circinata. Plant Dis 96:908
Swett CL, Gordon TR (2015) Endophytic association of the pine pathogen Fusarium circinatum with corn (Zea mays). Fungal Ecol 13:120–129
Swinton J, Gilligan CA (1999) Selecting hyperparasites for biocontrol of Dutch elm disease. Proc R Soc B 266:437–445
Szucs M, Eigenbrode SD, Schwarzlander M, Schaffner U (2012) Hybrid vigor in the biological control agent, Longitarsus jacobaeae. Evol Applic 5:489–497
Tainter FH, Baker FA (1996) Principles of forest pathology. John Wiley and Sons, Inc, New York, 803 pp
Takamatsu S, Braun U, Limkaisang S, Kom-un S, Sato Y, Cunnington JH (2007) Phylogeny and taxonomy of the oak powdery mildew Erysiphe alphitoides sensu lato. Mycol Res 111:809–826
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32
Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D (2006) Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philos T Roy Soc B 361:1947–1963
Tedersoo L, Bahram M, Põlme S, Yorou NS, Wijesundera R, Villarreal Ruiz L, Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Põldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Pärtel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson KH, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo LD, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, De Kesel A, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K (2014) Global diversity and geography of soil fungi. Science 346:1256688
Telford A, Cavers S, Ennos RA, Cottrell JE (2015) Can we protect forests by harnessing variation in resistance to pests and pathogens? Forestry 88:3–12
Tittonell P (2014) Ecological intensification—sustainable by nature. Curr Opin Environ Sustain 8:53–61
Tollenaere C, Pernechele B, Mäkinen HS, Parratt SR, Németh MZ, Kovács GM, Kiss L, Tack AJ, Laine AL (2014) A hyperparasite affects the population dynamics of a wild plant pathogen. Mol Ecol 23:5877–5887
Tsui CK-M, Farfan L, Roe AD, Rice AV, Cooke JEK, El-Kassaby YA, Hamelin RC (2014) Population structure of mountain pine beetle symbiont Leptographium longiclavatum and the implication on the multipartite beetle–fungi relationships. PLoS One 9, e105455
Turner TR, James EK, Poole PS (2013) The plant microbiome. Genome Biol 14:209
Vacher C, Daudin J-J, Piou D, Desprez-Loustau M-L (2010) Ecological integration of alien species into a tree–fungus network. Biol Invasions 12:3249–3259
Van der Most P, de Jong B, Parmentier HK, Verhulst S (2011) Trade-off between growth and immune function: a meta-analysis of selection experiments. Funct Ecol 25:74–80
van Schie CC, Takken FL (2014) Susceptibility genes: how to be a good host. Annu Rev Phytopathol 52:551–581
Vander Wal E, Garant D, Calmé S, Chapman C, Festa-Bianchet M, Millien V, Rioux-Paquette S, Pelletier F (2014) Applying evolutionary concepts to wildlife disease ecology and management. Evol Appl 7:856–868
Varki A (2012) Nothing in medicine makes sense, except in the light of evolution. J Mol Med (Berl) 90:481–494
Vayssier-Taussat M, Albina E, Citti C, Cosson J-F, Jacques M-A, Lebrun M-H, Le Loir Y, Ogliastro M, Petit MA, Roumagnac P, Candresse T (2014) Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front Cell Infect Microbiol 5:4–29
Verhoeven KJF, Biere A, Harvey JA, Putten WH (2009) Plant invaders and their novel natural enemies: who is naive? Ecol Lett 12:107–117
Vleeshouwers VG, Rietman H, Krenek P, Champouret N, Young C, Oh SK, Wang M, Bouwmeester K, Vosman B, Visser RG, Jacobsen E, Govers F, Kamoun S, Van der Vossen EA (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS One 3, e2875
Vogan PJ, Schoettle AW (2015) Selection for resistance to white pine blister rust affects the abiotic stress tolerances of limber pine. For Ecol Manage 344:110–119
Voth PD, Mairura L, Lockhart BE, May G (2006) Phylogeography of Ustilago maydis virus H1 in the USA and Mexico. J Gen Virol 87:3433–3441
Vuillaume F, Thebaud G, Urbino C, Forfert N, Granier M, Froissart R, Blanc S, Peterschmitt M (2011) Distribution of the phenotypic effects of random homologous recombination between two virus species. Plos Pathogens 7, e1002028. doi:10.1371/journal.ppat.1002028
Wainhouse D (2005) Biological control. In: Wainhouse D (ed) Ecological methods in forest pest management. Oxford University Press, Oxford, pp 109–126
Walters DR, Avrova A, Bingham IJ, Burnett FJ, Fountaine J, Havis ND, Hoad SP, Hughes G, Loosely M, Oxley SJP, Renwick A, Topp CFE, Newton AC (2012) Control of foliar diseases in barley: towards an integrated approach. Eur J Plant Pathol 133:33–73
Whitham TG, Bailey JK, Schweitzer JA, Shuster SM, Bangert RK, LeRoy CJ, Lonsdorf E, Allan GJ, DiFazio SP, Potts BM, Fischer DG, Gehring CA, Lindroth RL, Marks J, Hart SC, Wimp GM, Wooley SC (2006) A framework for community and ecosystem genetics: from genes to ecosystems. Nat Rev Genet 7:510–523
Williamson M, Fitter A (1996) The varying success of invaders. Ecology 77:1661–1665
Witzell J, Martín JA, Blumenstein K (2014) Ecological aspects of endophyte-based biocontrol of forest diseases. In: Verma VC, Gange AC (eds) Advances in endophytic research. Springer India, New Delhi, pp 321–333
Woods A, Coates KD, Hamann A (2005) Is an unprecedented Dothistroma needle blight epidemic related to climate change? Bioscience 55:761–769
Xhaard C, Barrès B, Andrieux A, Bousset L, Halkett F, Frey P (2012) Disentangling the genetic origins of a plant pathogen during disease spread using an original molecular epidemiology approach. Mol Ecol 21:2383–2398
Xie J, Jiang D (2014) New Insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu Rev Phytopathol 52:45–68
Yanchuk A, Allard G (2009) Tree improvement programmes for forest health—can they keep pace with climate changes? Unasylva 231:50–56
Yang C, Crowley DE, Borneman J, Keen NT (2001) Microbial phyllosphere populations are more complex than previously realized. Proc Natl Acad Sci U S A 98:3889–3894
Zhang DY, Sun GJ, Jiang XH (1999) Donald’s ideotype and growth redundancy: a game theoretical analysis. Field Crops Res 61:179–187
Zhao YJ, Hosoya T, Baral HO, Hosoka K, Kakishima M (2012) Hymenoscyphus pseudoalbidus, the correct name for Lambertella albida reported from Japan. Mycotaxon 122:25–41
Acknowledgments
The authors are very grateful to Catherine Bastien, Isabelle Goldringer, and François Lefevre for stimulating discussions, and to Sylvain Delzon for helpful comments.
Funding
BJ was funded by a grant from the French Ministry of Research and Education (MENRT no. 2011/AF/57).
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Jean-Michel Leban
Contribution of the co-authors
J. Aguayo, C. Husson, and B. Marçais wrote the first draft of the part dealing with fungal diversity, C. Dutech of the part dealing with pathogen evolution, M.-L. Desprez-Loustau and K. Hayden of the part on disease resistance, C. Robin of the part on hyperparasitism, C. Vacher and B. Jakushkin of the part on the tree microbiote, M. L. Desprez-Loustau of the introduction and conclusion. D. Piou, M. L. Desprez-Loustau and B. Marçais conceived Fig. 1. All co-authors contributed to the final writing and revision of the manuscript, with a special contribution of K. Hayden. Marie-Laure Desprez-Loustau initiated and coordinated the review.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Desprez-Loustau, ML., Aguayo, J., Dutech, C. et al. An evolutionary ecology perspective to address forest pathology challenges of today and tomorrow. Annals of Forest Science 73, 45–67 (2016). https://doi.org/10.1007/s13595-015-0487-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13595-015-0487-4