Abstract
The genetic basis of host resistance to parasites is a fundamental aspect of host-parasite co-evolution, yet the precise mechanisms often remain unclear. Here, we follow on from a previous study on the genetically mediated variation in resistance to two common fungal brood parasites that cause chalkbrood and stonebrood in the honey bee. We assessed whether genetically mediated variation in resistance can be explained by the baseline immunocompetence of different larval genotypes by correlating the constitutive expression of two key immune genes with the observed level of resistance of each larval genotype to four different fungal brood parasites. We found significant variation between larval genotypes in the constitutive expression of abaecin but not defensin 2, but despite a suggestion of negative correlations between gene expression and resistance level in older larvae, there was no consistent evidence that baseline abaecin expression is a relevant predictor of resistance to these parasites. These results suggest that the constitutive expression of abaecin appears to have a genetic basis in honey bee larvae but that mechanisms other than innate expression of antimicrobial peptides might be more important in defence against the specific fungal brood parasites assessed here.
Similar content being viewed by others
1 Introduction
The interactions between hosts and their parasites are a major driver of evolutionary adaptation (Anderson and May 1982). Due to their close evolutionary partnerships, the mechanisms involved in a host’s response to infection by a parasite are often genetically mediated (Carius et al. 2001; Agrawal and Lively 2002). There is accumulating evidence that this genetically mediated response to infection is a driver of the high levels of multiple mating (polyandry) witnessed in many social insect taxa (Brown and Schmid-Hempel 2003). For example, higher levels of within-colony genetic variation have been correlated with increased resistance to disease in ants (Hughes and Boomsma 2004, 2006; Reber et al. 2008), bumblebees (Baer and Schmid-Hempel 1999) and honey bees (Tarpy 2003; Tarpy and Seeley 2006).
Despite the general consensus that the genetic variation introduced into social insect colonies through polyandry facilitates disease resistance, the specific mechanisms remain unclear. Genetically based resistance mechanisms could stem from differences in individual immunity but in social species could also be a product of group-level ‘social immunity’ (Cremer et al. 2007; Wilson-Rich et al. 2009). The most effective mechanisms employed in resistance are likely to involve a combination of both mechanisms but will also be influenced by the biology of the specific parasite in question. The baseline level of immunocompetence of an individual, however, will impact on individual disease resistance, should social immune mechanisms fail (Boomsma et al. 2005). Such individual defences include cellular immune responses such as phagocytosis and encapsulation, and humoral immune responses such as the production of antimicrobial peptides (Schmid-Hempel 2005; Evans et al. 2006).
There is accumulating evidence that bees (and possibly Hymenoptera) have a reduced number of immune genes in comparison to other insects (Evans et al. 2006; Barribeau et al. 2015), but honey bees are nonetheless subject to a broad range of microparasites against which individual immune responses are effective (Morse and Flottum 1997) and which will therefore play an important role in reducing levels of parasites within the colony. The antimicrobial peptide (AMP) abaecin is a key honey bee immune effector involved in the response to infection by multiple parasites (Casteels et al. 1990; Evans and Pettis 2005; Evans et al. 2006) and has been shown to have significant heritable variation in its expression (Decanini et al. 2007). Additionally, another AMP shown to have considerable variation in expression after infection is defensin (Evans 2004), despite its expression levels not being associated with colony-level disease resistance (Evans and Pettis 2005). However, defensin takes two forms: defensin 1, which has been suggested to be involved in social immunity because it may potentially be transferred trophallactically, and defensin 2, which is more likely be involved in individual immunity (Klaudiny et al. 2005; Ilyasov et al. 2013).
Here, we follow on from a previous study showing the differential resistance of genotypes (patrilines) within colonies of honey bees to a set of fungal brood parasites (Evison et al. 2013), by assessing whether the variation in resistance levels of genotypes from one colony can be explained by differences in constitutive expression levels of the AMP genes abaecin and defensin 2. Considering honey bee AMPs are activated by the Toll pathway, which is stimulated by both bacteria and fungi (Evans et al. 2006), and both abaecin and defensin 1 have previously been shown to be upregulated in response to infection by A. apis (Aronstein et al. 2010), we hypothesised that AMPs may be involved in larval resistance to the fungi tested in Evison et al. (2013). First, we examine the level of variation in the constitutive expression of the two AMPs between different larval host genotypes in both 2nd and 5th instar larvae. Second, we correlate these expression levels with levels of resistance to two common brood parasites: the fungi Ascosphaera apis and Aspergillus flavus (the causative agents of chalkbrood and stonebrood disease, respectively). We were particularly interested in the variation in the response by different host genotypes to their obligate parasite A. apis (Chorbinski 2004), which is bound in a closely antagonistic host-parasite relationship, and therefore, we examined the response to three different strains of this parasite. For comparison, we also examined the host responses to a single strain of the ubiquitous fungus A. flavus that is an opportunistic parasite and is unlikely to have co-evolved with honey bees (Foley et al. 2012, 2014).
2 Material and methods
The level of resistance to four fungal parasites by the genotypes within a single colony of the European honey bee Apis mellifera carnica was first assessed through a survival experiment (Sect. 1), and host genotypes (patrilines) were assigned through microsatellite genotyping (Sect. 2; both detailed in Evison et al. 2013). Following this, the same host colony was used to assess the general level of immunocompetence of each host genotype through measuring expression of abaecin and defensin 2 in unchallenged larvae (Sect. 3). Finally, to assess whether this baseline immunocompetence level of each larval genotype explained their level of resistance to each parasite, the two measures were correlated (Sect. 4).
2.1 Assessment of resistance to fungal brood parasites
Individual variability in resistance to four fungal parasites (three strains of A. apis and one strain of A. flavus) was assessed through monitoring survival after controlled exposure to parasites. A total of 1440 larvae were reared individually in 48-well tissue culture plates on a diet of 50 % royal jelly, 6 % d-glucose, 6 % d-fructose and sterile deionised water, following a modified version of the procedures described by Aupinel et al. (2005) and Jensen et al. (2009). One- to 2-day-old larvae were removed from the comb using a Swiss grafting tool (Swinty, Sønderborg, Denmark) and transferred onto a droplet of larval diet within a cell culture plate. The plates were then placed in sealed boxes containing a pool of 0.04 % K2SO4 in order to establish high relative humidity and maintained at 34 °C. Larvae were fed daily ad libitum until they began to defecate (after moulting to the fifth instar); the wells were then cleaned with a cotton bud. Spores were harvested from media plates (Sabouraud dextrose agar) of each of the parasites. Each A. apis strain was formed by the mating of complementary mating type hyphae grown from two single-spore isolates. Strain E is formed by isolates KVL 07-098 + 06 117 and strain F by isolates KVL 06 123 + 06 132, all of which were obtained from the Fungal Entomopathogenic culture collection at the University of Copenhagen, Denmark (Vojvodic et al. 2011). Strain I was formed by isolates ARSEF 7405 + 7406, and the A. flavus strain was NRRL 1957, both of which were obtained from the Agricultural Research Service Entomopathogenic Fungus Collection, USA. Spore concentrations were prepared to a predetermined LD50 as follows: A. apis strain I 5.0 × 105, A. apis strain E 3.75 × 106, A. apis strain F 1.95 × 106 and A. flavus 1.0 × 105 spores/ml to account for differences between parasites in virulence and spore viability (which were determined as detailed in Evison et al. 2013; Vojvodic et al. 2011). The four parasite treatments plus the control treatment were administered to equal numbers of larvae (288 larvae dosed per treatment). Spore suspensions were applied directly to the mouth of larvae in 5-μL doses 2 days after grafting (or 5 μL sterilised water in the case of control larvae), and mortality and evidence of infection (hyphal growth) were monitored daily using a stereo microscope for 9 days following inoculation.
2.2 Genotyping of larvae
To assess the genetic basis of larval resistance to the three parasites, those larvae that died from infection by the parasites as well as those larvae that survived the 9-day period after inoculation were genotyped. Larvae that died due to other causes (which was 23 % of those under experimental conditions) showed rapid bacterial decomposition, which made them unsuitable for DNA extraction and were therefore excluded. A total of 698 larvae were successfully genotyped at eight microsatellite loci: A7, A29, B124, A35, A79, A107, A014 (Estoup et al. 1994) and AP243 (Solignac et al. 2003), with detailed protocols described in Evison et al. (2013). Multi-locus offspring genotypes were used to deduce the genotype of the colony queen and her multiple mates, and the workers were assigned to patrilines within the colony with extremely low detection errors (0.0001 %; Boomsma and Ratnieks 1996).
2.3 Gene expression
To gain insight into the immune mechanisms involved in resistance to these parasites, the expression levels of two key immune genes encoding for the antimicrobial peptides abaecin and defensin 2 were measured in another batch of larvae from the same colony. Larvae that had experienced the same rearing environment as those assessed for their level of resistance to each parasite but were never exposed to any of the parasites (83 full grown fifth instar larvae and 98 small second instar larvae) were collected and stored in RNA later at −20 °C for genotyping as above in combination with gene expression analysis. Total RNA was extracted from 5–10 mg of individual larval tissue using the Qiagen RNeasy Plus Mini Kit. RT-qPCR reactions were carried out independently for the immune genes abaecin and defensin 2 and for an endogenous control, the housekeeping gene RPS5, using 2 μL of RNA extract, 900 nM each primer and 250-nM TaqMan MGB Probes and the TaqMan Fast Virus 1-Step Master Mix in 10-μL reactions. Primers and probes for abaecin and RPS5 were as in Evans and Pettis (2005); the primers (D2F-CATTTCTGCAACTACCGCCTTT, D2R-CATTCAGCTTGCGCTATCAGAT) and probe (CGTCGTTGAGCTAAA) for defensin 2 were designed using Primer Express v3.0 (ABI) and the Apis mellifera defensin 2 sequence (NCBI ID number NM_001011638.1). Standard curve analyses were run for each assay, and the calculated efficiencies were 103 % for abaecin, 97 % for RPS5 and 98 % for defensin 2 over a 10,000-fold range. Reactions were run in a StepOne Plus instrument (ABI) in a Fast RT-PCR method of 50 °C for 5 min and 95 °C for 20 s followed by 40 cycles at 93 °C for 3 s and 60 °C for 30 s. Fluorescence data were collected each cycle during the extension step at 60 °C, and the data were analysed using the StepOne Software v2.1 (ABI) in a comparative C T (∆∆C T ) experiment using ROX as a passive reference signal to allow for the normalisation of the reporter dye signals. All reactions were carried out in triplicate (nine reactions per sample). Negative controls and a reference sample (calibrator) were also run in triplicate on each plate, and negative RT reactions were set up using the same Master Mix after incubation at 95 °C for 1 min (to allow for the inactivation of the retrotranscriptase) in order to confirm the absence of genomic DNA in the RNA extracts. To account for differences in the amount of starting material, ∆C T values were calculated by subtracting the mean C T value for RSP5 from the mean C T values for abaecin and defensin 2 in each sample. ∆∆C T values were then obtained by subtracting the ∆C T value in the reference sample from the ∆C T value for each gene in each sample. This ∆∆C T was then scaled as a power of 2 to obtain the estimate of relative cDNA abundances (RQ values).
2.4 Statistical analysis
All analysis was carried out using R statistical software (R Development Core Team 2014). Differences in survival of larvae between treatments and patrilines, and their interaction were analysed using Cox-proportional hazard survival models implemented using the coxph function of the survival package (Therneau 2011), with survivors of the experiment incorporated as right-censored data. The hazard ratio was extracted for each of the 13 patrilines as compared to the control survival and was calculated for the survival response to each of the four parasite treatments. The gene expression data was trimmed so that only patrilines with six or more individual larvae were represented, which reduced the number of patrilines analysed from 13 to eight (Table I). Differences between patrilines in gene expression were then assessed using a linear model of the log transformed expression data as the response variable, against patriline and age, and their interaction. Correlations between log transformed hazard ratio values and levels of log transformed gene expression in both old and young larvae were made using Pearson’s product-moment correlations, with a sequential Bonferroni correction applied post hoc to control for the multiple tests employed per gene/parasite/larval age combination.
3 Results
3.1 Effect of genotype on resistance to fungal brood parasites
There were significant differences in the survival of larvae treated with the four different parasites (χ 2 3 = 10.6; P = 0.014), and in overall survival between different genotypes of larvae (χ 2 12 = 36.9; P < 0.001), as well as an interaction between the two effects (χ 2 36 = 71.8; P < 0.001), indicating differences in resistance to the four parasite treatments between larval genotypes (Evison et al. 2013). Larvae showed genotypic differences in survival to all three strains of the A. apis parasite (E: χ 2 12 = 35.3; P < 0.001; F: χ 2 10 = 20.4; P = 0.026; I: χ 2 10 = 45.2; P < 0.001), but not to A. flavus (χ 2 11 = 12.3; P = 0.341). Control larvae survived well with minimal mortality (~19 %).
3.2 Effect of larval genotype on levels of gene expression
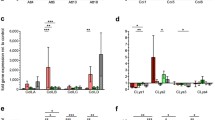
There was a significant effect of both age and larval genotype on levels of expression of abaecin (age: F 1,110 = 346.4, P < 0.001; patriline: F 6,110 = 10.7, P < 0.001), as well as a significant interaction between the two (F 6,110 = 4.79, P = 0.001), showing that expression levels differed between host genotypes as the larvae aged. Young larvae had negligible expression of abaecin, while expression in old larvae was highly variable, with patrilines varying as much as 4-fold (Figure 1a). Conversely, we found a significant effect of age on levels of expression of defensin 2 (age: F 1,110 = 14.1, P < 0.001; Figure 1b), but no effect of patriline (F 6,110 = 0.286, P = 0.942) and no interaction between the two (F 6, 110 = 0.348, P = 0.909). Both young and old larvae had moderate levels of expression of defensin 2, with this generally being slightly lower in young larvae and little difference between the host genotypes (Figure 1b).
The expression levels (relative quantity) of abaecin (a) and defensin 2 (b) in the seven genotypes (patrilines) of young (second instar—grey bars) and old (fifth instar—white bars) larvae found within the host colony. Y-axis of graph a is Log10 to enable assessment of the large difference in expression levels of abaecin between larval age groups.
3.3 Relationship between genotypic differences in resistance and immunocompetence
Correlations between the relative resistance of the larval genotypes (as measured by hazard ratio, HR) to each of the four parasites and the relative gene expression of either young or old larvae of each genotype appeared to show negative relationships for the older larvae but were not as clear for younger larvae (Figure 2). However, there was only one significant correlation found across all the possible 16 relationships after Bonferroni adjustment. This was between the responses by larvae to A. apis strain E and expression of abaecin (R = −0.418; t 63 = −3.65; P < 0.001; Figure 2c), with the relationship showing a negative correlation, i.e. a higher level of expression of abaecin in old larvae signified a higher level of resistance to strain E of A. apis.
Patterns of correlation between expression levels (relative quantity) of abaecin (a, c) and defensin 2 (b, d) shown by young (second instar, a, b) and old (fifth instar, c, d) larvae, and the levels of resistance (hazard ratio of exposure) shown to four parasite treatments by each of the seven genotypes within the host colony. White circles and solid grey line = A. flavus. Grey triangles and dashed black line = A. apis strain E. White triangles and dashed grey line = A. apis strain F. Grey circles and solid black line = A. apis strain I. The single significant correlation is highlighted with a thickened line (within c the responses by larvae to A. apis strain E and expression of abaecin; R = −0.418; t 63 = −3.654; P < 0.001).
4 Discussion
Here, we investigated the genetic basis to expression of two antimicrobial peptides (AMPs) involved in the humoral immunity of honey bees and whether this is likely to be a potential mechanism driving any genetically mediated resistance to the fungal brood parasites that cause chalkbrood and stonebrood disease (Invernizzi et al. 2009; Evison et al. 2013). In unchallenged larvae, we found a significant difference in the constitutive expression of the key immune gene abaecin between larval host genotypes but no genotypic variation in expression of another immune gene defensin 2. This suggests that levels of the AMP abaecin, but not defensin 2, might act as relevant predictor of host resistance to fungal brood parasites. For older larvae in particular, there appeared to be a trend for negative correlations between the expression levels of these two immune genes in different genotypes and their relative resistance to the three strains of A. apis parasite (i.e. that higher constitutive expression of these genes in unchallenged larvae led to a lower hazard ratio when challenged with the parasite). Those negative trends were less consistent for the A. flavus parasite, something we expected considering that we found a lack of genotypic variation in resistance to this parasite due to its looser evolutionary history with the honey bee (Evison et al. 2013). However, despite these trends, there was limited statistical support for any relationship between AMP expression and resistance, as only one relationship was significant and the slope was relatively shallow.
Despite the limited statistical support for a correlation between resistance by larval genotypes and our measures of their baseline immunocompetence, as well as the study only using larvae from a single colony, the patterns of constitutive expression of both abaecin and defensin 2 appear to be comparable to previous studies of these genes in response to another brood disease: American foulbrood disease (AFB), caused by the bacterial parasite Paenibacillus larvae (Evans 2004; Evans and Pettis 2005). Evans (2004) showed similar levels to our study in the constitutive expression of defensin as larvae aged. The AMP defensin takes two forms, defensin 1 and defensin 2, which are thought to have different roles within the honey bee colony. Defensin 1 appears to be associated with social immunity, and defensin 2 individual immunity (Klaudiny et al. 2005; Ilyasov et al. 2013). Our study specifically measured defensin 2 because we were interested in individual immunocompetence of larvae, but we found no significant variation in its expression between larval genotypes. Instead, it appeared to be relatively consistently expressed across genotypes in both young and old larvae. Defensin 2 is synthesised by the cells of the fat body and is regulated by interaction between both the Toll and Imd pathways (Ilyasov et al. 2013), suggesting that perhaps its role may be involved in more broad spectrum aspects of immunity and growth of honey bee larvae if its constituent expression is relatively uniform across genotype and ontogeny. This was in contrast to the expression levels of abaecin, which was expressed at a much higher level in old larvae, and its expression significantly differed between genotypes. We found much higher (and much more variable) constitutive levels of abaecin expression in older larvae compared to Evans (2004), even though our larvae that were measured for gene expression were never challenged with any parasites. This may simply be a product of the in vitro rearing conditions of the larvae, as abaecin production is thought to be affected by multiple genes with complex interactions (Decanini et al. 2007). However, its production may also have been induced by non-pathogenic (or even pathogenic) bacteria present in the laboratory setting or the guts of the larvae (Evans and Lopez 2004). Regardless of the reason for its expression in the absence of fungal challenge, and considering all larvae underwent the same rearing protocol, its significant variation between larval genotypes is interesting and suggests that its expression is under significant selection from parasites (Decanini et al. 2007).
The immune defences employed against fungi such as A. apis are similar to those employed in defence against bacteria such as P. larvae (Aronstein and Saldivar 2005; Ilyasov et al. 2013). Both fungi and gram-positive bacteria are known to induce the Toll signalling pathway in Drosophila melanogaster (Feldhaar and Gross 2008), and although honey bees have fewer functional AMPs than solitary insects such as D. melanogaster, the same pathways appear to be induced leading to upregulation of the same antimicrobial effectors, including abaecin, defensin 1 and defensin 2 (Evans et al. 2006). Evans and Pettis (2005) showed a strong negative relationship between colony levels of AFB and expression of abaecin in individual larvae, suggesting that higher expression of abaecin afforded more successful resistance against bacterial infection. However, they also showed a cost to the expression of abaecin inferring greater resistance, as colonies that mounted a stronger immune response to bacterial challenge also had lower fitness in terms of larval production. This suggests that even if constitutive expression levels of these AMPs vary significantly between host genotypes, expression should be minimised in the absence of parasites due to their cost of production (Evans and Pettis 2005). This might explain why we did not find a consistently strong relationship between baseline expression levels of abaecin and resistance levels of our larval genotypes, despite its significant variation in expression between patrilines. A paired design to measure the response in both challenged and unchallenged larval genotypes in multiple colonies, as well as tests of the direct effects of these AMPs on fungal growth using zone of inhibition assays, could provide further insight into these mechanisms.
Measuring the baseline immunocompetence of larvae by using measures of humoral defence mechanisms such as AMP levels should give a good representation of genetic variation in resistance by larvae to a variety of brood diseases. This is because larvae have limited ability to respond to or avoid parasites due to being confined to a cell during development; therefore, constitutive expression of innate immune mechanisms by some genotypes of larvae may afford greater protection for the colony when parasite pressure is high, through mechanisms such as herd immunity (Anderson and May 1985). However, because there appears to be complex regulation of AMPs in the honey bee, including potential cross-talk between different signalling pathways (Aronstein and Holloway 2013; Aronstein and Murray 2010), expression of single components of the immune mechanisms of the host may not provide the whole story. These complexities of the host’s immune defence introduce confounding factors in the patterns of specific host-parasite co-evolution. Furthermore, colony-level behavioural resistance mechanisms mediated by the workers such as hygienic behaviour (Invernizzi et al. 2011), resin collection (Simone-Finstrom and Spivak 2012), larval immunisation (Traniello et al. 2002) and larval gut microbiota (Omar et al. 2014) may represent a significant resistance pressure driving the patterns of parasite virulence and host resistance. Mechanisms such as these might uncouple the specific innate immune responses from resistance patterns witnessed within the colony.
Honey bee larvae also have other defence mechanisms to the parasites studied here, for example A. apis exposed larvae exhibit differential transcription of a chitinase-like enzyme which could be linked to anti-fungal activity in the gut (Aronstein et al. 2010). In addition, variation in honey bee resistance to A. apis appears to be based around single nucleotide polymorphisms, which are in close proximity to genes involved in the host’s chitin biosynthesis and development (Holloway et al. 2012), so the gut lining (its composition and/or its products) may be a key point of defence against infection. This may give further explanation as to why our measures of host baseline immunocompetence did not fully explain how different larval genotypes resist the brood parasites tested here. A key limitation of this study is that it is based on a single colony of honey bees and ideally should be expanded to assess between colony as well as within-colony variation in immunocompetence and resistance. Nevertheless, the apparent standing genetic variation in expression of abaecin corroborates previous studies on the importance of this immune effector in innate immune responses in honey bee larvae (Evans 2004; Evans and Pettis 2005; Decanini et al. 2007). This variation is likely to be important in resistance to the plethora of parasites honey bees are exposed to, often at the same time (Evans et al. 2006; Hedtke et al. 2011). Our results also suggest that the relationship between honey bees and their fungal brood parasites are extremely complex, probably due to the multiple interacting ecological and evolutionary factors driving this relationship (Evison et al. 2015).
References
Agrawal, A., Lively, C.M. (2002) Infection genetics: gene-for gene versus matching-alleles models and all points in between. Evol. Ecol. Res. 4, 79–90
Anderson, R.M., May, R.M. (1982) Coevolution of hosts and parasites. Parasitology 85, 411–426
Anderson, R.M., May, R.M. (1985) Vaccination and herd immunity to infectious diseases. Nature 318, 323–329
Aronstein KA, Holloway B (2013) Honey bee fungal pathogen, Ascosphaera apis; current understanding of host-pathogen interactions and host mechanisms of resistance. In Méndez-Vilas, A. (ed.) Microbial Pathogens and Strategies for Combating Them - Science, Technology and Education. Formatex Research Center, pp 402–410
Aronstein, K.A., Murray, K.D. (2010) Chalkbrood disease in honey bees. J. Invertebr. Pathol. 103, S20–S29
Aronstein, K.A., Saldivar, E. (2005) Characterization of a honeybee Toll related receptor gene Am18w and its potential involvement in antimicrobial immune defense. Apidologie 36, 3–14
Aronstein, K.A., Murray, K.D., Saldivar, E. (2010) Transcriptional responses in Honey bee larvae infected with chalkbrood fungus. BMC Genomics 11, 391
Aupinel, P., Fortini, D., Dufour, H., Tasei, J.-N., Michaud, B., Odoux, J.-F., et al. (2005) Improvement of artificial feeding in a standard in vitro method for rearing Apis mellifera larvae. Bull. Insectology 58, 107–111
Baer, B., Schmid-Hempel, P. (1999) Experimental variation in polyandry affects parasite load and fitness in a bumblebee. Nature 397, 151–154
Barribeau, S.M., Sadd, B.M., du Plessis, L., Brown, M.J.F., Buechel, S.D., et al. (2015) A depauperate immune repertoire precedes evolution of sociality in bees. Genome Biol. 16, 83
Boomsma, J.J., Ratnieks, F.L.W. (1996) Paternity in eusocial Hymenoptera. Philos. Trans. R. Soc. Lond. B 351, 947–975
Boomsma, J.J., Schmid-Hempel, P., Hughes, W.O.H. (2005) Life histories and parasite pressure across the major groups of social insects. In: Fellowes, M., Holloway, G., Rolff, J. (eds.) Insect evolutionary ecology, pp. 139–175. CABI, Wallingford
Brown, M.J.F., Schmid-Hempel, P. (2003) The evolution of female multiple mating in social Hymenoptera. Evolution 57, 2067–2081
Carius, H.J., Little, T.J., Ebert, D. (2001) Genetic variation in a host-parasite association: potential for coevolution and frequency-dependent selection. Evolution 55, 1136–1145
Casteels, P., Ampe, C., Riviere, L., Van Damme, J., Elicone, C., et al. (1990) Isolation and characterization of abaecin, a major antimicrobial response peptide in the honeybee (Apis mellifera). Eur. J. Biochem. 187, 381–386
Chorbinski, P. (2004) The development of the infection of Apis mellifera larvae by Ascosphaera apis. Elec. J. Pol. Agri. Univ. Vet. Med. 7
Cremer, S., Armitage, S.A.O., Schmid-Hempel, P. (2007) Social immunity. Curr. Biol. 17, R693–R702
Decanini, L.I., Collins, A.M., Evans, J.D. (2007) Variation and heritability in immune gene expression by diseased honeybees. J. Hered. 98, 195–201
Development Core Team, R. (2014) R: a language and environment for statistical computing. R foundation for Statistical Computing, Vienna
Estoup, A., Solignac, M., Cournet, J.M. (1994) Precise assessment of the number of patrilines and of genetic relatedness in honeybee colonies. Proc. R. Soc. Lond. B 258, 1–7
Evans, J.D. (2004) Transcriptional immune responses by honey bee larvae during invasion by the bacterial pathogen, Paenibacillus larvae. J. Invertebr. Pathol. 85, 105–111
Evans, J.D., Lopez, D.L. (2004) Bacterial probiotics induce an immune response in the honey bee (Hymenoptera: Apidae). J. Econ. Entomol. 97, 752–756
Evans, J.D., Pettis, J.S. (2005) Colony-level impacts of immune responsiveness in honey bees, Apis mellifera. Evolution 59, 2270–2274
Evans, J.D., Aronstein, K., Chen, Y., Hetru, C., Imler, J.-L., Jiang, H., Kanost, M., Thomspon, J.G., Zou, Z., Hultmark, D. (2006) Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 15, 645–656
Evison, S.E.F., Fazio, G., Chappell, P., Foley, K., Jensen, A.B., Hughes, W.O.H. (2013) Host-parasite genotypic interactions in the honey bee: the dynamics of diversity. Ecol. Evol. 3, 2214–2222
Evison, S.E.F., Foley, K., Jensen, A.B., Hughes, W.O.H. (2015) Genetic diversity, virulence and fitness evolution in an obligate fungal parasite of bees. J. Evol. Biol. 28, 179–188
Feldhaar, H., Gross, R. (2008) Immune reactions of insects on bacterial pathogens and mutualists. Microbes Infect. 10, 1082–1088
Foley, K., Fazio, G., Jensen, A., Hughes, W.O.H. (2012) Nutritional limitation and resistance to opportunistic Aspergillus parasites in honey bee larvae. J. Invertebr. Pathol. 111, 68–73
Foley, K., Fazio, G., Jensen, A.B., Hughes, W.O.H. (2014) The distribution of Aspergillus spp. opportunistic parasites in hives and their pathogenicity to honey bees. Vet. Microbiol. 169, 203–210
Hedtke, K., Jensen, A.B., Genersch, E. (2011) Evidence for emerging parasites and pathogens influencing outbreaks of stress-related diseases like chalkbrood. J. Invertebr. Pathol. 108, 167–173
Holloway, B., Sylvester, H.A., Bourgeois, L., Rinderer, T.E. (2012) Association of single nucleotide polymorphisms to resistance to chalkbrood in Apis mellifera. J Apic. Res. 51, 154–163
Hughes, W.O.H., Boomsma, J.J. (2004) Genetic diversity and disease resistance in leaf-cutting ant societies. Evolution 58, 1251–1260
Hughes, W.O.H., Boomsma, J.J. (2006) Does genetic diversity hinder parasite evolution in social insect colonies? J. Evol. Biol. 19, 132–143
Ilyasov, R.A., Gaifullina, L.R., Saltykova, E.S., Poskryakov, A.V., Nikolaenko, A.G. (2013) Defensins in the honeybee antiinfectious protection. J. Evol. Biochem. Physiol. 49, 1–9
Invernizzi, C., Peñagaricano, F., Tomasco, I.H. (2009) Intracolonial genetic variability in honeybee larval resistance to the chalkbrood and American foulbrood parasites. Insectes Soc. 56, 233–240
Invernizzi, C., Rivas, F., Bettucci, L. (2011) Resistance to chalkbrood disease in Apis mellifera L. (Hymenoptera: Apidae) colonies with different hygienic behaviour. Neotrop. Entomol. 40, 28–34
Jensen, A.B., Pedersen, B.V.B., Eilenberg, J. (2009) Differential susceptibility across honey bee colonies in larval chalkbrood resistance. Apidologie 40, 524–534
Klaudiny, J., Albert, S., Bachanová, K., Kopernicky, J., Simúth, J. (2005) Two structurally different defensin genes, one of them encoding a novel defensin isoform, are expressed in honeybee Apis mellifera. Insect Biochem. Mol. Biol. 35, 11–22
Morse, R.A., Flottum, K. (1997) Honey bee pests, predators, and diseases. The A.I. Root Co., OH
Omar, M.O.M., Moustafa, A.M., Ansari, M.J., Anwar, Fahmy, B.F., Al-Ghamdi, A., Nuru, A. (2014) Antagonistic effect of gut bacteria in the hybrid carnolan honey bee Apis mellifera carnica, against Ascosphaera apis, the causal organism of chalkbrood disease. J. Apic. Sci. 58, 17–27
Reber, A., Castella, G., Christe, P., Chapuisat, M. (2008) Experimentally increased group diversity improves disease resistance in an ant species. Ecol. Lett. 11, 682–689
Schmid-Hempel, P. (2005) Evolutionary ecology of insect immune defences. Annu. Rev. Entomol. 50, 529–551
Simone-Finstrom, M.D., Spivak, M. (2012) Increased resin collection after parasite challenge: a case of self-medication in honey bees? PLoS One 7, e34601
Solignac, M., Vautrin, D., Loiseau, A., Mougel, F., Baudry, E., Estoup, A., et al. (2003) Five hundred and fifty microsatellite markers for the study of the honeybee (Apis mellifera L.) genome. Mol. Ecol. Notes 3, 307–311
Tarpy, D.R. (2003) Genetic diversity within honeybee colonies prevents severe infections and promotes colony growth. Proc. R. Soc. Lond. B 270, 99–103
Tarpy, D.R., Seeley, T.D. (2006) Lower disease infections in honeybee (Apis mellifera) colonies headed by polyandrous vs monandrous queens. Naturwissenschaften 93, 195–199
Therneau, T.M. (2011) Package ‘survival’. Available at http://cran.r-project.org/web/packages/survival/survival.pdf (accessed March 09, 2011).
Traniello, J.F.A., Rosengaus, R.B., Savoi, K. (2002) The development of immunity in a social insect: evidence for the group facilitation of disease resistance. Proc. Natl. Acad. Sci. U.S.A. 99, 6838–6842
Vojvodic, S., Jensen, A.B., Markussen, B., Eilenberg, J., Boomsma, J.J. (2011) Genetic variation in virulence among chalkbrood strains infecting honeybees. PLoS ONE 6, e25035
Wilson-Rich, N., Spivak, M., Fefferman, N.H., Starks, P.T. (2009) Genetic, individual, and group facilitation of disease resistance in insect societies. Annu. Rev. Entomol. 54, 405–423
Acknowledgments
We thank Bill Cadmore for apicultural assistance, three anonymous reviewers and members of the Hughes Lab for comments on the work, Svjetlana Vojvodic and Sofie Schmidt for technical assistance, and the Natural Environment Research Council for funding (NE/G006849/1). We also thank the Agricultural Research Service of the United States Department of Agriculture for providing the Aspergillus flavus strain NRRL 1957.
Author information
Authors and Affiliations
Corresponding author
Additional information
Manuscript Editor: Stan Schneider
L'expression des peptides antimicrobiens dans la réponse immunitaire innée n'explique pas la diversité génotypique dans la résistance aux champignons parasites du couvain chez l'abeille
Apis mellifera / Ascosphaera apis / Aspergillus flavus / abaecine/ défensine
Die Expression antimikrobieller Peptide in der angeborenen Immunantwort erklärt nicht die genetische Diversität in der Resistenz der Honigbienenbrut gegen parasitische Pilze
Apis mellifera / Abaecin / Defensin / antimikrobielle Peptide / Ascosphaera apis / Aspergillus flavus
Rights and permissions
About this article
Cite this article
Evison, S.E.F., Fazio, G., Chappell, P. et al. Innate expression of antimicrobial peptides does not explain genotypic diversity in resistance to fungal brood parasites in the honey bee. Apidologie 47, 206–215 (2016). https://doi.org/10.1007/s13592-015-0388-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-015-0388-4