Abstract
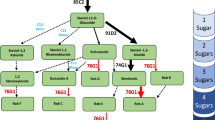
Stevia plants are well-known for their ability to synthesize steviol glycosides (SGs), a natural sweetener blend. The principal SGs include stevioside (STV) and Rebaudioside-A (Reb-A), with the latter exhibiting superior sweetness and organoleptic properties. UDP glucosyltransferase-76G1 (UGT76G1) is responsible for converting STV to Reb-A, determining the intensity of sweetness. A better understanding of the structure/activity of SrUGT76G1 could provide insights into Reb-A production in stevia plants. To this end, a combination of enzymatic assays and sequencing analysis was performed using two stevia genotypes (Brazilian and Spanish) with contrasting Reb-A production capabilities (off/on). Relative expression of SrUGT76G1 gene showed remarkably higher expression (~ threefold) in Spanish samples compared to Brazilian ones. Foliar protein fractions (crude or partially purified extract) from Brazil plants were unable to convert STV into Reb-A under in vitro conditions, resulting in undetectable levels of Reb-A by HPLC. Molecular analyses revealed that the Brazilian SrUGT76G1 gene not only presents a premature stop codon, resulting in the absence of PSPG motif responsible for the binding of glycosyl groups, but also exhibits mutations affecting key amino acid residues in the acceptor-binding pocket. These alterations provide a plausible explanation for the Brazilian protein inability to catalyze the transformation of STV into Reb-A.
Graphical abstract







Similar content being viewed by others
Abbreviations
- GGDP:
-
Geranylgeranyl diphosphate
- KA:
-
Kaurenoic acid
- MEP:
-
2-C-methyl-D-erythritol-4 phosphate
- NCBI:
-
National Center for Biotechnology Information
- PSPG:
-
Plant secondary product glycosyltransferase
- Reb-A:
-
Rebaudioside-A
- Reb-B:
-
Rebaudioside-B
- Reb-D:
-
Rebaudioside-D
- Reb-E:
-
Rebaudioside-E
- Reb-G:
-
Rebaudioside-G
- Reb-I:
-
Rebaudioside-I
- Reb-M:
-
Rebaudioside-M
- Reb-Q:
-
Rebaudioside-Q
- SGs:
-
Steviol glycosides
- SrUGT:
-
Uridine-diphosphate glycosyltransferase in Stevia rebaudiana
- STV:
-
Stevioside
- UGT:
-
Uridine-diphosphate glycosyltransferase
- UGT73E1:
-
UDP glucosyltransferase-73E1
- UGT74G1:
-
UDP glucosyltransferase-74G1
- UGT76G1:
-
UDP glucosyltransferase-76G1
- UGT85C2:
-
UDP glucosyltransferase-85C2
- UGT91D2:
-
UDP glucosyltransferase-91D2
References
Abdelsalam NR, Botros WA, Khaled AE, Ghonema MA, Hussein SG, Ali HM, Elshikh MS (2019) Comparison of uridine diphosphate-glycosyltransferase UGT76G1 genes from some varieties of Stevia rebaudiana Bertoni. Sci Rep 9:8559. https://doi.org/10.1038/s41598-019-44989-4
Ahmad J, Khan I, Blundell R, Azzopardi J, Mahomoodally MF (2020) Stevia rebaudiana Bertoni.: an updated review of its health benefits, industrial applications and safety. Trends Food Sci Tech 100:177–189. https://doi.org/10.1016/j.tifs.2020.04.030
Álvarez-Robles MJ, López-Orenes A, Ferrer MA, Calderón AA (2016) Methanol elicits the accumulation of bioactive steviol glycosides and phenolics in Stevia rebaudiana shoot cultures. Ind Crops Prod 87:273–279. https://doi.org/10.1016/j.indcrop.2016.04.054
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. https://doi.org/10.1093/bioinformatics/bti770
Bogado-Villalba L, Nakayama Nakashima H, Britos R et al (2021) Genotypic characterization and steviol glycoside quantification in a population of Stevia rebaudiana Bertoni from Paraguay. J Crop Sci Biotechnol 24:145–152. https://doi.org/10.1007/s12892-020-00066-1
Brandle JE, Telmer PG (2007) Steviol glycoside biosynthesis. Phytochemistry 68:1855–1863. https://doi.org/10.1016/j.phytochem.2007.02.010
Buchan DW, Ward S, Lobley AE, Nugent T, Bryson K, Jones DT (2010) Protein annotation and modelling servers at University College London. Nucl Acids Res 38:563–568. https://doi.org/10.1093/nar/gkq427
Ceunen S, Geuns JMC (2013) Steviol glycosides: chemical diversity, metabolism, and function. J Nat Prod 76:1201–1228. https://doi.org/10.1021/np400203b
Chughtai MFJ, Pashab I, Zahoor T, Khaliq A, Ahsan S, Wuc Z, Nadeem M, Mehmood T, Amire RM, Yasminf I, Liaqat A, Tanweerh S (2020) Nutritional and therapeutic perspectives of Stevia rebaudiana as emerging sweetener; a way forward for sweetener industry. Food 18:164–177. https://doi.org/10.1080/19476337.2020.1721562
de Andrade MVS, Lucho SR, do Amaral MN, et al (2023) Long-day photoperiodic requirements for steviol glycosides and gibberellins biosynthesis and bio-sweetener levels optimization in Stevia rebaudiana Bertoni. Ind Crops Prod 204:117363. https://doi.org/10.1016/j.indcrop.2023.117363
Dyduch-Siemińska M, Najda A, Gawroński J et al (2020) Stevia rebaudiana Bertoni, a source of high-potency natural sweetener—biochemical and genetic characterization. Molecules 25:767. https://doi.org/10.3390/molecules25040767
Gerwig GJ, Poele EM, Dijkhuizen L, Kamerling LP (2017) Structural analysis of rebaudioside a derivatives obtained by Lactobacillus reuteri 180 glucansucrase-catalyzed trans-α-glucosylation. Carbohydr Res 440:51–62. https://doi.org/10.1016/j.carres.2017.01.008
Geuns JMC (2003) Stevioside. Phytochemistry 64:913–921
Hall A (1999) Bioedit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hernández KV, Moreno-Romero J, Hernández de la Torre M et al (2022) Effect of light intensity on steviol glycosides production in leaves of Stevia rebaudiana plants. Phytochemistry 194:113027. https://doi.org/10.1016/j.phytochem.2021.113027
JECFA (2010) Steviol glycosides. In: Compendium of food additive specifications. FAO JECFA Monographs vol 10, pp. 17–21.
Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202. https://doi.org/10.1006/jmbi.1999.3091
Kim MJ, Zheng J, Liao MH, Jang IC (2019) Overexpression of SrUGT76G1 in Stevia alters major steviol glycosides composition towards improved quality. Plant Biotechnol J 17:1037–1047. https://doi.org/10.1111/pbi.13035
Kumar H, Kaul K, Bajpai-Gupta S, Kaul VK, Kumar S (2012) A comprehensive analysis of fifteen genes of steviol glycosides biosynthesis pathway in Stevia rebaudiana (Bertoni). Gene 492:276–284. https://doi.org/10.1016/j.gene.2011.10.015
Lee SG, Salomona E, Yud O, Jeza JM (2019) Molecular basis for branched steviol glucoside biosynthesis. PNAS 116:13131–13136. https://doi.org/10.1073/pnas.1902104116
Lemus-Mondaca R, Vega-Gálvez A, Zura-Bravo L, Ah-Hen K (2012) Stevia rebaudiana Bertoni, source of a high-potency natural sweetener: a comprehensive review on the biochemical, nutritional and functional aspects. Food Chem 132:1121–1132. https://doi.org/10.1016/j.foodchem.2011.11.140
Li W, Zhou Y, You W et al (2018) Development of photoaffinity probe for the discovery of steviol glycosides biosynthesis pathway in Stevia rebaudiana and rapid substrate screening. ACS Chem Biol 13:1944–1949. https://doi.org/10.1021/acschembio8b00285
Liu Z, Li J, Sun Y, Zhang P, Wang Y (2020) Structural insights into the catalytic mechanism of a plant diterpene glycosyltransferase SrUGT76G1. Plant Commun 1:100004. https://doi.org/10.1016/j.xplc.2019.100004
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lucho SR, do Amaral MR, Auller PA, Bianchi VJ, Ferrer MA, Calderón AA, Braga EJB, (2019) Salt stress-induced changes in in vitro cultured Stevia rebaudiana Bertoni: effect on metabolite contents, antioxidant capacity and expression of steviol glycosides-related biosynthetic genes. J Plant Growth Regul 38:1341–1353. https://doi.org/10.1007/s00344-019-09937-6
Lucho SR, Amaral MN, Benitez LC, Milech C, Kleinowski AM, Bianchi VJ, Braga EJB (2018) Validation of reference genes for RT-qPCR studies in Stevia rebaudiana in response to elicitor agents. Physiol Mol Biol Plants 24:767–779. https://doi.org/10.1007/s12298-018-0583-7
Lucho SR, do Amaral MN, Milech C, Ferrer MA, Calderón AA, Bianchi VJ, Braga EJB (2018) Elicitor-induced transcriptional changes of genes of the steviol glycosides biosynthesis pathway in Stevia rebaudiana Bertoni. J Plant Growth Regul 37:971–985. https://doi.org/10.1007/s00344-018-9795-x
Lucho SR, do Amaral MN, Milech C, Bianchi VJ, Almagro L, Ferrer MA, Calderón AA, Braga EJB (2021) Gibberellin reverses the negative effect of paclobutrazol but not of chlorocholine chloride on the expression of SGs/GAs biosynthesis-related genes and increases the levels of relevant metabolites in Stevia rebaudiana. Plant Cell Tiss Organ Cult 146:171–184. https://doi.org/10.1007/s11240-021-02059-6
Madhav H, Bhasker S, Chinnamma M (2012) Functional and structural variation of uridine diphosphate glycosyltransferase (UGT) gene of Stevia rebaudiana–UGTSr involved in the synthesis of rebaudioside A. Plant Physiol Biochem 63:245–253. https://doi.org/10.1016/j.plaphy.2012.11.029
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–479
Olsson K, Carlsen S, Semmler A, Simón E, Mikkelsen MD, Moller BL (2016) Microbial production of next-generation stevia sweeteners. Microb Cell Fact 15:207. https://doi.org/10.1186/s12934-016-0609-1
Petit E, Jacques A, Daydé J, Vallejo V, Berger M (2019) UGT76G1 polymorphism in Stevia rebaudiana: new variants for steviol glycosides conjugation as revealed by expression analysis of regulatory genes and metabolic inhibitors studies. Plant Physiol Biochem 135:563–569. https://doi.org/10.1016/j.plaphy.2018.11.002
Richman AS, Gijzen M, Starratt AN, Yang Z, Brandle JE (1999) Diterpene synthesis in Stevia rebaudiana: recruitment and up-regulation of key enzymes from the gibberellin biosynthetic pathway. Plant J 19:411–421
Richman A, Swanson A, Humphrey T, Chapman R, Mcgarvey B, Pocs R, Brandle J (2005) Functional genomics uncovers three glucosyltransferases involved in the synthesis of the major sweet glucosides of Stevia rebaudiana. Plant J 41:56–67. https://doi.org/10.1111/j.1365-313X.2004.02275.x
Saifi M, Yogindran S, Nasrullah N, Nissar U, Gul I, Abdin MZ (2019) Co-expression of anti-miR319g and miRStv_11 lead to enhanced steviol glycosides content in Stevia rebaudiana. BMC Plant Biol 19:274. https://doi.org/10.1186/s12870-019-1871-2
Ullah A, Munir S, Mabkhot Y, Badshah S (2019) Bioactivity profile of the diterpene isosteviol and its derivatives. Molecules 24:678. https://doi.org/10.3390/molecules24040678
Wang J, Li S, Xiong Z, Wang Y (2016) Pathway mining-based integration of critical enzyme parts for de novo biosynthesis of steviolglycosides sweetener in Escherichia coli. Cell Res 26:258–261. https://doi.org/10.1038/cr.2015.111
Wu Q, La Hovary C, Chen H-Y et al (2020) An efficient Stevia rebaudiana transformation system and in vitro enzyme assays reveal novel insights into UGT76G1 function. Sci Rep 10:3773. https://doi.org/10.1038/s41598-020-60776-y
Yadav SK, Guleria P (2012) Steviol glycosides from Stevia: biosynthesis pathway review and their application in foods and medicine. Crit Rev Food Sci Nutr 52:988–998. https://doi.org/10.1080/10408398.2010.519447
Yang Y, Huang S, Han Y, Yuan H, Gu S, Zhao Y (2014) Base substitution mutations in uridinediphosphate-dependent glycosyltransferase 76G1 gene of Stevia rebaudiana causes the low levels of rebaudioside a Mutations in UGT76G1, A key gene of steviol glycosides synthesis. Plant Physiol Biochem 80:220–225. https://doi.org/10.1016/j.plaphy.2014.04.005
Yang Y, Huang S, Han Y et al (2015) Environmental cues induce changes of steviol glycosides contents and transcription of corresponding biosynthetic genes in Stevia rebaudiana. Plant Physiol Biochem 86:174–180. https://doi.org/10.1016/j.plaphy.2014.12.004
Zhang S, Chen H, Xiao J, Liu Q, Xiao R, Wu W (2019) Mutations in the uridine diphosphate glucosyltransferase 76G1 gene result in different contents of the major steviol glycosides in Stevia rebaudiana. Phytochemistry 162:141–147. https://doi.org/10.1016/j.phytochem.2019.03.008
Zhang S, Liu Q, Lyu C, Chen J, Xiao R, Chen J, Yang Y, Zhang H, Hou K, Wu W (2020) Characterizing glycosyltransferases by a combination of sequencing platforms applied to the leaf tissues of Stevia rebaudiana. BMC Genom 21:794. https://doi.org/10.1186/s12864-020-07195-5
Acknowledgements
The authors gratefully acknowledge the CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) for their financial support and research fellowship EJBB and VJB, as well as the FAPERGS (Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul). This study was financed in part by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior Brasil—Finance Code 001). We are also grateful to the Structural Genomics Laboratory belonging to the Technological Development Center (CDTec) of the Federal University of Pelotas (UFPEL). This work was partially carried out at IBV (Instituto de Biotecnología Vegetal), UPCT (Spain).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13562_2024_888_MOESM1_ESM.docx
Supplementary file1: Amplified SrUGT76G1 gene in Stevia rebaudiana. Lane 1: 1kb DNA ladder; lane 2: DNA band from Brazil; lane 3: DNA band from Spain; lane 4: cDNA from Brazil; lane 5: cDNA from Spain, and lane 6: NC-Negative Control. (DOCX 14 kb)
13562_2024_888_MOESM2_ESM.pdf
Supplementary file2: Sequence plot of SrUGT76G1 proteins predicted by PSIPRED. (A) Reference (AY345974.1), (B) Spain, and (C) Brazil (PDF 109 kb)
13562_2024_888_MOESM5_ESM.docx
Supplementary file5: Similarity, identity, and gap data among SrUGT76G1 sequences from Brazil (accession number MZ781502.1), Spain (accession number MZ781503.1) and Reference (accession number AY345974.1). (DOCX 15 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lucho, S.R., do Amaral, M.N., Bianchi, V.J. et al. Sequencing analysis and enzyme activity assay of SrUGT76G1 revealed the mechanism toward on/off production of Rebaudioside-A in stevia plants. J. Plant Biochem. Biotechnol. (2024). https://doi.org/10.1007/s13562-024-00888-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13562-024-00888-y