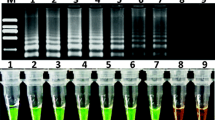
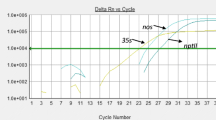
Abstract
Genetically modified (GM) food crops for desired traits have been approved in some of the countries. The approval status of a GM event varies from country to country. In India, Bt cotton is the only approved GM crop. So far, no GM food crop has got regulatory approval in the country, which may be considered as unauthorized GM (UGM) event in the Indian context. The entry of UGM events of food crops needs to be checked in the imported consignments as well as in the marketplace. In the present study, screening elements were identified based on the genetically modified organism (GMO) matrix developed as decision support system for 22 GM food crops approved globally. Three sets of multiplex PCR assays were developed and validated for GM detection in food crops and products: triplex PCR targeting control elements [Cauliflower Mosaic Virus 35S promoter (P-35S), Figwort Mosaic Virus promoter (P-FMV), nopaline synthase terminator (T-nos)], triplex PCR targeting marker genes [aminoglycoside-3’-adenyltransferase (aadA), neomycin phosphotransferase (nptII), phosphinothricin-N-acetyltransferase (pat)] and duplex PCR targeting Bt genes (cry1Ab/Ac and cry2Ab2). Limit of detection (LOD) ranged from 0.5–0.01% for different targets, which is in compliance with the labelling threshold of many countries. The developed assays were utilized to check the GM status of apple and maize products along with an additional test for ctp2-cp4epsps for herbicide tolerance in maize. Based on the tests conducted, none of these samples were found positive for GM targets screened. These procedures could be efficiently employed as a part of GMO testing to trace GM contamination, if any, in both the imported as well as domestic food products.





Similar content being viewed by others
Data availability Sstatement
Data sharing is not applicable to this article as no datasets were generated or analyzed. All the data is included and provided in the main text of the manuscript or in the Supplementary Information.
Abbreviations
- aadA :
-
Aminoglycoside-3′-adenyltransferase
- cp4-epsps :
-
5-Enol pyruvylshikimate-3-phosphate synthase
- CRM:
-
Certified reference material
- CTAB:
-
Cetyltrimethyl ammonium bromide
- DNA:
-
Deoxyribonucleic acid
- EU:
-
European Union
- FSSAI:
-
Food Safety and Standards Authority of India
- GM:
-
Genetically modified
- GMO:
-
Genetically modified organism
- LOD:
-
Limit of detection
- nptII :
-
Neomycin phosphotransferase
- P-35S :
-
Cauliflower Mosaic Virus 35S promoter
- pat :
-
Phosphinothricin-N-acetyltransferase
- PCR:
-
Polymerase chain reaction
- P-FMV :
-
Figwort Mosaic Virus promoter
- RM:
-
Reference material
- T-nos :
-
Nopaline synthase terminator
- UGM:
-
Unauthorized genetically modified
References
Avsar B, Sadeghi S, Turkec A, Lucas SJ (2020) Identification and quantitation of genetically modified (GM) ingredients in maize, rice, soybean and wheat-containing retail foods and feeds in Turkey. J Food Sci Technol 57(2):787–793. https://doi.org/10.1007/s13197-019-04080-2
Bak A, Emerson JB (2019) Multiplex quantitative PCR for single reaction genetically modified (GM) plant detection and identification of false positive GM plants linked to Cauliflower mosaic virus (CaMV) infection. BMC Biotechnol 19:73. https://doi.org/10.1186/s12896-019-0571-1
Cardarelli P, Branquinho MR, Ferreira RTB, da Cruz FP, Gemal AL (2005) Detection of GMO in food products in Brazil: the INCQS experience. Food Control 16(10):859–866. https://doi.org/10.1016/j.foodcont.2004.07.010
Datukishvili N, Kutateladze T, Gabriadze I, Bitskinashvili K, Vishnepolsky B (2015) New multiplex PCR methods for rapid screening of genetically modified organisms in foods. Front Microbiol 6:757. https://doi.org/10.3389/fmicb.2015.00757
Eum SJ, Kim IR, Lim HS, Lee JR, Choi W (2019) Event-specific multiplex PCR method for four genetically modified cotton varieties, and its application. Appl Biol Chem 62:52. https://doi.org/10.1186/s13765-019-0459-8
FSSA (2006) The Food Safety and Standards Act, 2006. https://www.indiacode.nic.in/bitstream/123456789/7800/1/200634_food_safety_and_standards_act%2C_2006.pdf. Accessed May 4, 2022
FSSAI (2021) https://fssai.gov.in/upload/advisories/2021/02/60212749e94b1Clarification_GM_Food_08_02_2021.pdf. Accessed May 4, 2022
Grohmann L, Brunen Nieweler C, Nemeth A, Waiblinger HU (2009) Collaborative trial validation studies of real-time PCR based GMO screening methods for detection of the bar gene and the ctp2-cp4epsps construct. J Agric Food Chem 57:8913–8920. https://doi.org/10.1021/jf901598r
Grohmann L, Reiting R, Mäde D, Uhlig S, Simon K, Frost K, Randhawa GJ, Zur K (2015) Collaborative trial validation of cry1Ab/Ac and Pubi-cry TaqMan-based real-time PCR assays for detection of DNA derived from genetically modified Bt plant products. Accred Qual Assur 20:85–96. https://doi.org/10.1007/s00769-015-1108-5
Holden MJ, Levine M, Scholdberg T, Haynes RJ, Ronald JG (2010) The use of 35S and Tnos expression elements in the measurement of genetically engineered plant materials. Anal Bioanal Chem 396:2175–2187. https://doi.org/10.1007/s00216-009-3186-x
Holst-Jensen A, Bertheau Y, de Loose M, Grohmann L, Hamels S, Hougs L, Morisset D, Pecoraro S, Pla M, Van den Bulcke M, Wulff D (2012) Detecting unauthorized genetically modified organisms (GMOs) and derived materials. Biotechnol Adv 30(6):1318–1335. https://doi.org/10.1016/j.biotechadv.2012.01.024
http://bch.cbd.int/database/organisms. Accessed Feb 23, 2022
http://www.isaaa.org/gmapprovaldatabase/. Accessed May 22, 2022
https://ec.europa.eu/jrc/en/research-topic/gmos. Accessed Jan 6, 2022
https://geacindia.gov.in/meetings.aspx. Accessed May 22, 2022
ISO/TS 21569-5:2016 (2016) Horizontal methods for molecular biomarker analysis—methods of analysis for the detection of genetically modified organisms and derived products—part 5: real-time PCR based screening method for the detection of the FMV promoter (P-FMV) DNA sequence
ISO21569:1-69 (2005) Foodstuffs—methods of analysis for the detection of genetically modified organisms and derived products—qualitative nucleic acid based methods
James D, Schmidt AM, Wall E, Green M, Masri S (2003) Reliable detection and identification of genetically modified maize, soybean, and canola by multiplex PCR analysis. J Agric Food Chem 51(20):5829–5834. https://doi.org/10.1021/jf0341159
JRC Technical Report (2015) Definition of minimum performance requirements for analytical methods of GMO testing. European Network of GMO Laboratories (ENGL), European Commission—Joint Research Centre. Ispra (VA), Italy
Kim JH, Jeong D, Kim YR, Kwon YK, Rhee GS, Zhang D (2013) Development of a multiplex PCR method for testing six GM soybean events. Food Control 31:366–371. https://doi.org/10.1016/j.foodcont.2012.10.015
Kralj Novak P, Gruden K, Morisset D, Lavrac N, Stebih D, Rotter A, Zel J (2009) GMOtrack: generator of cost-effective GMO testing strategies. J AOAC Int 92:1739–1746 (PMID: 20166592)
Lipp M, Bluth A, Eyquem F, Kruse L, Schimmel H, Van den Eede G, Anklam E (2001) Validation of a method based on polymerase chain reaction for the detection of genetically modified organisms in various processed foodstuffs. Eur Food Res Technol 212:497–504. https://doi.org/10.1007/s002170000274
Lipp M, Broadmann P, Pietsch K, Pauwels J, Anklam E (1999) IUPAC collaborative trial study of a method to detect genetically modified soybeans and maize in dried powder. J AOAC Int 82(4):923–928. https://doi.org/10.1093/jaoac/82.4.923
Mazzara M, Grazioli E, Savini C, Van Den Eede G (2013) Event-specific method for the quantification of maize line T25 using real-time PCR v. 1.01—validation report and validated method. Online Publication
Permingeat HR, Reggiardo MI, Vallejos RH (2002) Detection and quantification of transgenes in grains by multiplex and real time PCR. J Agric Food Chem 50:4431–4436. https://doi.org/10.1021/jf020081d
Querci M, Van den Bulcke M, Žel J, Van den Eede G, Broll H (2010) New approaches in GMO detection. Anal Bioanal Chem 6:1991–2002. https://doi.org/10.1007/s00216-009-3237-3
Ramos-Gómez S, Busto MD, Perez-Mateos M, Ortega N (2014) Development of a method to recovery and amplification DNA by real-time PCR from commercial vegetable oils. Food Chem 158:374–383. https://doi.org/10.1016/j.foodchem.2014.02.142
Randhawa GJ, Chhabra R, Singh M (2009) Multiplex PCR based simultaneous amplification of selectable marker and reporter genes for the screening of genetically modified crops. J Agric Food Chem 57:5167–5172. https://doi.org/10.1021/jf900604h
Randhawa GJ, Chhabra R, Singh M (2010) Decaplex and real time PCR-based detection of MON531 and MON15985 Bt cotton events. J Agric Food Chem 58(18):9875–9881. https://doi.org/10.1021/jf100466n
Randhawa GJ, Morisset D, Singh M, Žel J (2014) GMO matrix: a cost-effective approach for screening for unauthorized genetically modified events in India. Food Control 38:124–129. https://doi.org/10.1016/j.foodcont.2013.10.013
Randhawa GJ, Singh M, Sood P (2016) DNA-based methods for detection of genetically modified events in food and supply chain. Curr Sci 110(6):1000–1009. https://doi.org/10.18520/cs/v110/i6/1000-1009
Rantakokko-Jalava K, Jalava J (2002) Optimal DNA isolation method for detection of bacteria in clinical specimens by broad-range PCR. J Clin Microbiol 40(11):4211–4217. https://doi.org/10.1128/JCM.40.11.4211-4217.2002
Reischl U, Linde HJ, Metz M, Leppmeier B, Lehn N (2000) Rapid identification of methicillin-resistant Staphylococcus aureus and simultaneous species confirmation using real-time fluorescence PCR. J Clin Microbiol 38(6):2429–2433. https://doi.org/10.1128/JCM.38.6.2429-2433.2000
Singh M, Randhawa GJ (2016) Transboundary movement of genetically modified organisms in India: Current scenario and a decision support system. Food Control 68:20–24. https://doi.org/10.1016/j.foodcont.2016.03.032
Singh M, Bhoge RK, Randhawa G (2016) Crop-specific GMO matrix-multiplex PCR: a cost-efficient screening strategy for genetically modified maize and cotton events approved globally. Food Control 70:271–280. https://doi.org/10.1016/j.foodcont.2016.05.032
Singh M, Sodhi KK, Paliwal A, Sharma S, Randhawa G (2021) Efficient DNA extraction procedures for processed food derivatives—a critical step to ensure quality for GMO analysis. Food Anal Methods 14:2249–2261. https://doi.org/10.1007/s12161-021-02051-y
Singh M, Kaur K, Sharma S, Paliwal A, Singh M, Aminedi R, Kaur V, Randhawa G (2022) Assessment of risk of GM contamination in flaxseed accessions imported from Canada: a case study to restrict the unauthorized GM events from entering India. Plant Genet Resou: Char & Utiliz (in press)
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mole Biol 17(5):1105–1109. https://doi.org/10.1007/BF00037152
Van den Bulcke M, Lievens A, Barbau-Piednoir E, Mbongolombella G, Roosens N, Sneyers M, Casi AL (2010) A theoretical introduction to “combinatory SYBRGreen qPCR screening”, a matrix-based approach for the detection of materials derived from genetically modified plants. Anal Bioanal Chem 396(6):2113–2123. https://doi.org/10.1007/s00216-009-3286-7
Van Tongeren SP, Degener JE, Harmsen HJM (2011) Comparison of three rapid and easy bacterial DNA extraction methods for use with quantitative real-time PCR. Eur J Clin Microbiol Infec Dis 30(9):1053–1061. https://doi.org/10.1007/s10096-011-1191-4
Waiblinger HU, Grohmann L, Mankertz J, Engelbert D, Pietsch K (2010) A practical approach to screen for authorised and unauthorised genetically modified plants. Anal Bioanal Chem 396(6):2065–2072. https://doi.org/10.1007/s00216-009-3173-2
Acknowledgements
The authors thank the Indian Council of Agricultural Research (ICAR) and the Director, ICAR-National Bureau of Plant Genetic Resources for providing necessary facilities and resources.
Funding
The grant provided by the Department of Biotechnology (DBT) is duly acknowledged.
Author information
Authors and Affiliations
Contributions
MS: conceptualization, supervision, data curation, formal analysis, writing original draft; drafting of figures and tables; AP, KK: methodology, validation; PP: data analysis and compilation for GMO matrix; GR: administration, supervision, review of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Disclaimer
Any opinions, findings, or conclusions, expressed in this publication are those of the authors and do not necessarily reflect the view of the concerned organization.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, M., Paliwal, A., Kaur, K. et al. Development and utilization of analytical methods for rapid GM detection in processed food products: a case study for regulatory requirement. J. Plant Biochem. Biotechnol. 32, 511–524 (2023). https://doi.org/10.1007/s13562-023-00832-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-023-00832-6