Abstract
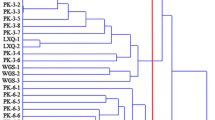
Okra is an important vegetable crop that provides a significant portion of vitamins and minerals for populations in several countries. Okra has been cultivated in Turkey for centuries, and was likely introduced by the Arabs from Africa in ancient times. In this study, we aimed to clarify the genetic variation within 35 Turkish okra germplasm, by comparing it against 25 different genotypes from India, Africa, and the United States, using 30 morphological characters and 19 sequence-related amplified polymorphism (SRAP) primer combinations. Fruit, leaf, and stem color were the primary characteristics to distinguish the okra accessions. Those features, among 30 individual phenotypic traits, explained 42 % of phenotypic variation in the first three axes of the principal component analysis (PCA) with leaf shape, flower size, cotyledon length, fruit-surface angularity, cotyledon width, and petal color. Phenotypic observation results showed that while 1051 Togo (10.76), Red Wonder (7.99), TR-05-1 (7.2), 1159 Togo (7.17), and Red Balady (7.15) were found to be more divergent accessions, Cajun Queen (5.06), Perkins Spineless (5.09), Jade (5.18), TR-01-1 (5.2), and DLGG (5.32) were the closest okra accessions. According to phenotypic data, Turkish okra accessions were located adjacent to the Indian, American, and African okra accessions in clusters three and four. However, marker data showed that African okra possessed a more distinct form compared to the other okra germplasms. Nineteen SRAP primer combinations produced 92 bands and 29 (31.5 %) of them were found to be polymorphic among okra accessions. 1051 Togo was found to be the most divergent accession in phenotypic observation.





Similar content being viewed by others
Abbreviations
- PIC:
-
Polymorphism information contents
- UPGMA:
-
The Unweighted Pair-Group Method with Arithmetic Mean
- PCA:
-
The principal component analysis
References
Aladele SE, Ariyo OJ, Lapena R (2008) Genetic relationship among West African okra (Abelmoschus caillei) and Asian genotypes (Abelmoschus esculentus) using RAPD. Afr J Biotechnol 7:1426–1431
Ariyo OJ (1987) Multivariate analysis and the choice of parents for hybridization in okra (Abelmoschus esculentus (L.) Moench). Theor Appl Genet 74:361–363
Ariyo OJ, Odulaja A (1991) Numerical analysis of variation among accessions of okra (Abelmoschus esculentus (L.) Moench), Malvaceae. Ann Bot 67:527–531
Boiteux LS, Fonseca MEN, Simon PW (1999) Effects of plant tissue and DNA purification methods on randomly amplified polymorphic DNA-based genetic fingerprinting analysis in carrot. J Am Soc Hortic Sci 124:32–38
Corley WL (1985) UGA red okra: a new edible ornamental. Research report, University of Georgia, College of Agriculture, Experiment Stations, No: 484
Dash GB (1997) Multivariate analysis in okra (Abelmoschus esculentus (L.) Moench). Environ Ecol 15:332–334
Dhankhar BS, Mishra JP (2009) Origin, history, and distribution. In: Dhankhar BS, Singh R (eds) Okra handbook: Global production, processing, and crop improvement. HNB Publishing, New York, pp 3–23
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Düzyaman E (1997) Okra: botany and horticulture. Hortic Rev 21:41–72
Düzyaman E (2005) Phenotypic diversity within a collection of distinct okra (Abelmoschus esculentus) cultivars derived from Turkish land races. Genet Resour Crop Evol 52:1019–1030
Düzyaman E (2009) Okra in Turkey. In: Dhankhar BS, Singh R (eds) Okra handbook: Global production, processing, and crop improvement, HNB Publishing, New York, pp 323–346
Düzyaman E, Vural H (2002) Different approaches in the improvement process of local okra varieties. Acta Hortic 579:139–144
FAO (2014) http://faostat.fao.org/
Ferriol M, Pico B, Nuez F (2003) Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP markers. Theor Appl Genet 107:271–282
Gulsen O, Karagul S, Abak K (2007) Diversity and relationships among Turkish okra germplasm by SRAP and phenotypic marker polymorphism. Biol Bratislava 62:41–45
Hamon S, Charrier A (1983) Large variation of okra collected in Benin and Togo. Plant Genet Resour News Lett 56:52–58
Hamon S, Noirot M (1991) Some proposed procedures for obtaining a core collection using quantitative plant characterization data. International workshop on okra genetic resources held at the National Bureau for Plant Genetic Resources, 1990 International Crop Network Series 5:89–94
Kyriakopoulou OG, Arens P, Pelgrom KTB, Karapanos I, Bebeli P, Passam HC (2014) Genetic and morphological diversity of okra (Abelmoschus esculentus L. Moench.) genotypes and their possible relationships, with particular reference to Greek landraces. Sci Hortic 171:58–70
Mahajan RK, Bisht IS, Agrawal RC, Rana RS (1996) Studies on South Asian okra collection: methodology for establishing a representative core set using characterization data. Genet Resour Crop Evol 43:249–25
Martin FW, Rhodes AM, Ortiz M, Diaz F (1981) Variation in okra. Euphytica 30:697–705
Rohlf FJ (1997) NTSYs-Pc: numerical taxonomy and multivariate analysis system. Exeter Software, New Work
SAS Institute (2005) SAS online Doc, Version 8. SAS Inst., Cary, NC
Schafleitner R, Kumar S, Lin C, Hegde SG, Ebert A (2013) The okra (Abelmoschus esculentus) transcriptome as a source for gene sequence information and molecular markers for diversity analysis. Gene 517:27–36
Scott SJ, McFerran J, Goode MJ (1990) ‘Jade’ okra. HortSci 25(127):128
Seck A (1991) Okra germplasm evaluation in Senegal. Workshop on okra genetic resources. In Report of an international workshop on okra genetic resources held at the National Bureau for Plant Genetic Resources, International Crop Network Series N°5., Rome: IBPGR. New Delhi, India, 8–12 Oct. 1990
UPOV (1999) Guidelines for the conduct of tests for distinctness, uniformity and stability; Okra (Abelmoschus esculentus (L.) Moench.). TG/167/3 GENEVA
Vural H, Esiyok D, Duman I (2000) Cultivated vegetables (Vegetable production). Ege University Press, Bornova (in Turkish)
Weir BS (1990) Genetics data analysis: methods for discrete population genetic data. Sinaur Associates, Sunderland
Yildiz M, Ekbic E, Keles D, Sensoy S, Abak K (2011) Use of ISSR, SRAP, and RAPD markers to assess genetic diversity in Turkish melons. Sci Hortic 130:349–353
Acknowledgments
This work was supported by the Scientific Research Projects Unit of Cukurova University (BAP Project number: ZF2009 BAP18). We would like to express our gratitude to AARI (Aegean Agricultural Research Institute) for providing some seed samples. We would like to acknowledge Dr. Vanessa S. Gordon who improved the language of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yıldız, M., Ekbiç, E., Düzyaman, E. et al. Genetic and phenotypic variation of Turkish Okra (Abelmoschus esculentus L. Moench) accessions and their possible relationship with American, Indian and African germplasms. J. Plant Biochem. Biotechnol. 25, 234–244 (2016). https://doi.org/10.1007/s13562-015-0330-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-015-0330-x