Abstract
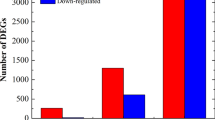
To better understand disease resistance genes in white poplars, a total of 74 resistance gene homologs (RGHs) with a nucleotide-binding site (NBS) were investigated from the Populus trichocarpa genome, and the expression patterns of these genes in various tissues of a triploid white poplar [(P. tomentosa × P. bolleana) × P. tomentosa] under different growth conditions were examined using quantitative real-time PCR. The results showed that 27 genes were expressed in six of the examined tissues of triploid poplar at various levels. Six, two and three genes were significantly more expressed in apical leaves, young bark and mature bark respectively, than in other tissues, suggesting a potential involvement in tissue specific disease resistance or other functions. Twenty-four genes had dramatically higher expression in apical leaves than in mature leaves, and 22 genes had a higher expression in mature bark than in young bark, suggesting developmental stage-specificity of gene expression in poplar tissues. In addition, wounding induced expression of 22 genes and reduced expression of three genes. Growth in dark conditions caused the up-regulation of 20 genes and the down-regulation of one gene. While methyl jasmonic acid (MeJA) and salicylic acid (SA) induced transcriptional changes of 19 genes, but only one gene was regulated in a simultaneous way. Treatment with the compatible Agrobacterium tumefaciens induced 11 genes. These data suggested that there are complex interconnecting signal transduction pathways regulating the expression of poplar NBS-encoding genes.



Similar content being viewed by others
References
Bakker E, Borm T, Prins P, Vossen EVD, Uenk G, Arens M, Boer JD, Eck HV, Muskens M, Vossen J, Linden GVD, Ham RV, Klein-lankhorst R, Visser R, Smant G, Bakker J, Goverse A (2011) A genome-wide genetic map of NB-LRR disease resistance loci in potato. Theor Appl Genet 123:493–508
Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94
Century KS, Lagman RA, Adkisson M, Morlan J, Tobias R, Schwartz K, Smith A, Love J, Ronald PC, Whalen MC (1999) Development control of Xa21-mediated disease resistance in rice. Plant J 20:231–236
Charrier B, Champion A, Henry Y, Kreis M (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol 130:577–590
Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, Hulbert S, Pryor T (1999) Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11:1365–1376
Constabel CP, Yip L, Patton JJ, Christopher ME (2000) Polyphenol oxidase from hybrid poplar: cloning and expression in response to wounding and herbivory. Plant Physiol 124:285–295
Dowkiw A, Bastien C (2004) Characterization of two major genetic factors controlling quantitative resistance to Melampsora larici-populina leaf rust in hybrid poplars: strain specificity, field expression, combined effects, and relationship with defeated qualitative resistance gene. Phytopathology 94:1358–1367
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95:14863–14868
Glowacki S, Macioszek VK, Kononowicz AK (2011) Cell Mol Biol Lett 16:1–24
Goff SA, Ricke D, La TH et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Graham MA, Marek LF, Shoemaker RC (2002) Organization, expression and evolution of a disease resistance gene cluster in soybean. Genetics 162:1961–1977
Hazen SP, Wu Y, Kreps JA (2003) Gene expression profiling of plant responses to abiotic stress. Funct Integr Genom 3:105–111
Kachroo A, Kachroo P (2007) Salicylic acid-, jasmonic acid—and ethylene-mediated regulation of plant defense signaling. Genet Eng 28:55–83
Kohler A, Rinaldi C, Duplessis S, Baucher M, Geelen D, Duchaussoy F, Meyers BC, Boerjan W, Martin F (2008) Genome-wide identification of NBS resistance genes in populus trichocarpa. Plant Mol Biol 66:619–636
Koornneef A, Pieterse CMM (2008) Cross talk in defense signaling. Plant Physiol 146:839–844
Kuang HH, Woo SS, Meyers BC, Nevo E, Michelmore RW (2004) Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in Lettuce. Plant Cell 16:2870–2894
Leon J, Rojo E, Sanchez-Serrano JJ (2001) Wound signaling in plants. J Exp Bot 52:1–9
Lin YZ, Zheng HQ, Zhang Q, Liu CX, Zhang ZY (2013) Functional profiling of EcaICE1 transcription factor gene from Eucalyptus camaldulensis involved in cold response in tobacco plants. J Plant Biochem Biotechnol. doi:10.1007/s13562-013-0192-z
Liu JJ, Ekramoddoullah AKM (2004) Isolation, genetic variation and expression of TIR-NBS-LRR resistance gene analogs from western white pine (Pinus monticola Dougl. Ex. D. Don.). Mol Genet Genomics 270:432–441
Liu JJ, Ekramoddoullah AKM (2011) Genomic organization, induced expression and promoter activity of a resistance gene analog (PmTNL1) in western white pine (Pinus monticola). Planta 233(5):1041–1053
Love AJ, Yun BW, Laval V, Loake GJ, Milner JJ (2005) Cauliflower mosaic virus, a compatible pathogen of Arabidopsis, engages three distinct defense-signaling pathways and activates rapid systemic generation of reactive oxygen species. Plant Physiol 139:935–948
Lupas A, Vandyke M, Stock J (1991) Predicting coiled-coils from protein sequences. Science 252:1162–1164
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR encoding genes in Arabidopsis. Plant Cell 15:809–834
Mun JH YUHJ, Park S, Park BS (2009) Genome-wide identification of NBS-encoding resistance genes in Brassica rapa. Mol Genet Genom 282:617–631
Radwan O, Mouzeyar S, Nicolas P, Bouzidi MF (2005) Induction of a sunflower CC-NBS-LRR resistance gene analogue during incompatible interaction with Plasmopara halstedii. J Exp Bot 56:567–575
Rinaldi C, Kohler A, Frey P, Duchaussoy F, Ningre N, Couloux A, Wincker P, Thiec DL, Fluch S, Martin F, Duplessis S (2007) Transcript profiling of poplar leaves upon infection with compatible and incompatible strains of the foliar rust Melampsora larici-populina. Plant Physiol 144:347–366
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shen KA, Chin DB, Arroyo-Garcia R, Ochoa OE, Lavelle DO, Wroblewski T, Meyers BC, Michelmore RW (2002) Dm3 is one member of a large constitutively expressed family of nucleotide binding site-leucine-rich repeat encoding genes. Mol Plant-Microbe Interact 15:251–261
Takahashi H, Kanayama Y, Zheng MS, Kusano T, Hase S, Ikegami M, Jyoti S (2004) Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to cucumber mosaic virus. Plant Cell Physiol 45:803–809
Tameling WIL, Elzinga SDJ, Darmin PS, Vossen JH, Takken FLW, Haring MA, Cornelissen BJC (2002) The tomato R gene products I-2 and Mi-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14:2929–2939
Tan X, Meyers BC, Kozik A, West MA, Morgante M, St Clair DA, Bent AF, Michelmore RW (2007) Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC Plant Biol 7:56
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Thurau T, Kifle S, Jung C, Cai D (2003) The promoter of the nematode resistance gene Hs1pro-1 activates a nematode-responsive and feeding site-specific gene expression in sugar beet (Beta vulgaris L.) and Arabidopsis thaliana. Plant Mol Biol 52:643–660
Tuskan GA, DiFazio S, Jansson S, et al. (2006) The genome of black cottonwood, populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Wang ZX, Yano M, Yamanouchi U, Iwamoto M, Monna L, Hayasaka H, Katayose Y, Sasaki T (1999) The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J 19:55–64
Wang ZX, Yamanouchi U, Katayose Y, Sasaki T, Yano M (2001) Expression of the Pib rice-blast-resistance gene family is up-regulated by environmental conditions favouring infection and by chemical signals that trigger secondary plant defenses. Plant Mol Biol 47:653–661
Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang ZX, Kono I, Kurata N, Yano M, Iwata N, Sasaki T (1998) Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc Natl Acad Sci U S A 95:1663–1668
Zhang ZY, Li FL, Zhu ZT (1992) Chromosome doubling and triploid breeding of Populus tomentosa Carr. and its hybrid. J Beijing For Univ 14:52–58
Zhang ZY, Li FL, Zhu ZT (1997) Doubling technology of pollen chromosome of Populus tomentosa and its hybrids. J Beijing For Univ 6:9–20
Zhang Q, Zhang ZY, Lin SZ, Lin YZ (2005) Resistance of transgenic hybrid triploids in populus tomentosa Carr. against 3 species of Lepidopterans following two winter dormancies conferred by high level expression of cowpea trypsin inhibitor gene. Silvae Genet 54:108–116
Zhang Q, Zhang ZY, Lin SZ, Zheng HQ, An XM, Li Y, Li HX (2008) Characterization of resistance gene analogs with a nucleotide binding site isolated from a triploid white poplar. Plant Biol 10:310–322
Zheng HQ, Lin SZ, Zhang Q, Lei Y, Zhang ZY (2009) Functional analysis of 5’ untranslated region of a TIR-NBS-encoding gene from triploid white poplar. Mol Genet Genomics 282:381–394
Zheng HQ, Lin SZ, Zhang Q, Lei Y, Hou L, Zhang ZY (2010) Functional identification and regulation of the PtDrl02 gene promoter from triploid white poplar. Plant Cell Rep 29:449–460
Zheng HQ, Zhang Q, Li HX, Lin SZ, An XM, Zhang ZY (2011) Over-expression of the triploid white poplar PtDrl01 gene in tobacco enhances resistance to tobacco mosaic virus. Plant Biol 13:145–153
Zheng HQ, Lei Y, Zhang ZY, Lin SZ, Zhang Q, Liu WF, Du J, An XM, Zhao ZY (2012) Analysis of promoter activity of PtDrlo2 gene in white poplars. J Plant Biochem Biotechnol 1:88–97
Acknowledgments
This research was supported by National Natural Science Foundation of China (Grant No. 31270698), by National TCM Project Application in the 11th Five-Year Period in China (Grant No. 2006BAD01A15), by the Doctorate Program of University of Ministry of Education (grant No. 20070022003), and by the National Project of Transgenic Plants and Their Applications in China (Grant No. J2002-B-003). We thank Rongling Wu and Arthur Berg at the University of Florida for their help in revising this manuscript. We also thank two anonymous reviewers for construction comments and suggestions which greatly improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, Q., Lin, S., Zheng, H. et al. Expression profiling of NBS-encoding genes in a triploid white poplar. J. Plant Biochem. Biotechnol. 24, 283–291 (2015). https://doi.org/10.1007/s13562-014-0270-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-014-0270-x