Abstract
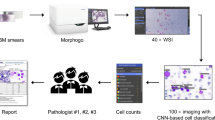
The detection, counting, and precise segmentation of white blood cells in cytological images are vital steps in the effective diagnosis of several cancers. This paper introduces an efficient method for automatic recognition of white blood cells in peripheral blood and bone marrow images based on deep learning to alleviate tedious tasks for hematologists in clinical practice. First, input image pre-processing was proposed before applying a deep neural network model adapted to cells localization and segmentation. Then, model outputs were improved by using combined predictions and corrections. Finally, a new algorithm that uses the cooperation between model results and spatial information was implemented to improve the segmentation quality. To implement our model, python language, Tensorflow, and Keras libraries were used. The calculations were executed using NVIDIA GPU 1080, while the datasets used in our experiments came from patients in the Hemobiology service of Tlemcen Hospital (Algeria). The results were promising and showed the efficiency, power, and speed of the proposed method compared to the state-of-the-art methods. In addition to its accuracy of 95.73%, the proposed approach provided fast predictions (less than 1 s).



Similar content being viewed by others
References
Naik S, Doyle S, Agner S, Madabhushi A, Feldman M, Tomaszewski J. Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology. In: 5th IEEE international symposium on biomedical imaging: from nano to macro, 2008. ISBI 2008. IEEE; 2008. p. 284–87.
Xu J, Xiang L, Liu Q, Gilmore H, Wu J, Tang J, Madabhushi A. Stacked sparse autoencoder (SSAE) for nuclei detection on breast cancer histopathology images. IEEE Trans Med Imaging. 2016;35(1):119–30.
Settouti N, Bechar MEA, Daho MEH, Chikh MA. An optimised pixel-based classification approach for automatic white blood cells segmentation. Int J Biomed Eng Technol. 2020;32(2):144–60.
Saidi M, Bechar MEA, Settouti N, Chikh MA. Instances selection algorithm by ensemble margin. J Exp Theor Artif Intell. 2018;30(3):457–78.
Benazzouz M, Baghli I, Benomar A, Ammar M, Benmouna Y, Chikh M. Evidential segmentation scheme of bone marrow images. Adv Image Video Process. 2016;4(1):37.
Baghli I, Nakib A, Sellam E, Benazzouz M, Chikh A, Petit E. Hybrid framework based on evidence theory for blood cell image segmentation. In: Medical imaging 2014: biomedical applications in molecular, structural, and functional imaging, vol. 9038. International Society for Optics and Photonics; 2014. p. 903815.
Benomar ML, Chikh MA, Descombes X, Benazzouz M. Multi features based approach for white blood cells segmentation and classification in peripheral blood and bone marrow images. Int J Biomed Eng Technol. 2019. https://doi.org/10.1504/IJBET.2019.10030162.
Zhang Z, Luo P, Loy CC, Tang X. Facial landmark detection by deep multi-task learning. In: European conference on computer vision. Springer; 2014. p. 94–108.
Dhungel N, Carneiro G, Bradley AP. Deep learning and structured prediction for the segmentation of mass in mammograms. In: International conference on medical image computing and computer-assisted intervention. Springer; 2015. p. 605–12.
Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer; 2015. p. 234–41.
Roth HR, Lu L, Farag A, Shin H-C, Liu J, Turkbey EB, Summers RM. Deeporgan: multi-level deep convolutional networks for automated pancreas segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer; 2015. p. 556–64.
Chen H, Dou Q, Ni D, Cheng J-Z, Qin J, Li S, Heng P-A. Automatic fetal ultrasound standard plane detection using knowledge transferred recurrent neural networks. In: International conference on medical image computing and computer-assisted intervention. Springer; 2015. p. 507–14.
Dou Q, Chen H, Jin Y, Yu L, Qin J, Heng P-A. 3d deeply supervised network for automatic liver segmentation from CT volumes. In: International conference on medical image computing and computer-assisted intervention. Springer; 2016. p. 149–57.
Dou Q, Chen H, Yu L, Zhao L, Qin J, Wang D, Mok VC, Shi L, Heng P-A. Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imaging. 2016;35(5):1182–95.
Żejmo M, Kowal M, Korbicz J, Monczak R. Classification of breast cancer cytological specimen using convolutional neural network. J Phys Conf Ser. 2017;783:012060.
Graham S, Vu QD, Raza SEA, Azam A, Tsang YW, Kwak JT, Rajpoot N. Hover-net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med Image Anal. 2019;58:101563.
Akram SU, Kannala J, Eklund L, Heikkilä J. Cell segmentation proposal network for microscopy image analysis. In: Deep learning and data labeling for medical applications. Springer; 2016. p. 21–9.
Dai J, He K, Sun J. Instance-aware semantic segmentation via multi-task network cascades. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016. p. 3150–8.
Yi J, Wu P, Huang Q, Qu H, Liu B, Hoeppner DJ, Metaxas DN. Multi-scale cell instance segmentation with keypoint graph based bounding boxes. In: International conference on medical image computing and computer-assisted intervention. Springer; 2019. p. 369–77.
He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. In: 2017 IEEE international conference on computer vision (ICCV). IEEE; 2017. p. 2980–8.
Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. In: Advances in neural information processing systems, 2015. p. 91–9.
Lin T-Y, Dollár P, Girshick R, He K, Hariharan B, Belongie S. Feature pyramid networks for object detection. In: CVPR, vol. 1; 2017, p. 4.
Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? In: Advances in neural information processing systems, 2014. p. 3320–8.
Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging. 2016;35(5):1299–312.
Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–98.
Prechelt L. Early Stopping — But When?. In: Neural Networks: Tricks of the Trade. Lecture Notes in Computer Science. Berlin: Springer; 2012. vol 7700, p. 53–67. https://doi.org/10.1007/978-3-642-35289-8_5.
Chen H, Qi X, Yu L, Dou Q, Qin J, Heng P-A. DCAN: deep contour-aware networks for object instance segmentation from histology images. Med Image Anal. 2017;36:135–46.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khouani, A., El Habib Daho, M., Mahmoudi, S.A. et al. Automated recognition of white blood cells using deep learning. Biomed. Eng. Lett. 10, 359–367 (2020). https://doi.org/10.1007/s13534-020-00168-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13534-020-00168-3