Abstract
Purpose
The objective of this paper is to present a generalized multiple classifier system for improved classification of mammographic masses in Computer-aided detection (CAD).
Methods
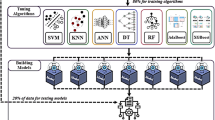
To encourage different base (component) classifiers to learn different parts of an object instant space, we develop a novel base classifier generation algorithm which combines data resampling underpinning AdaBoost with the use of different feature representations. In addition, our proposed multiple classifier system can be generalized beyond the limitation of weak classifiers in conventional AdaBoost learning. To this end, our multiple classifier system has an effective and efficient mechanism for tuning the level of weakness of base classifiers.
Results
Extensive experiments have been performed using benchmark mammogram data set to test the proposed method on classification between mammographic masses and normal tissues. In addition, to assess classification performance, we used the area under the receiver operating characteristic (AUC) and the normalized partial area under the curve (pAUC). Results show that our method considerably outperforms (in terms of both AUC and pAUC) the most commonly used single neural network (NN) and support vector machine (SVM) based classification approaches. In particular, the effectiveness of our method in terms of correct classification is much more significant over difficult mammogram cases with dense tissues that have higher risk of cancer incidences and cause higher false-positive (FP) detections.
Conclusions
Our multiple classifier system shows quite promising results in terms of improving classification performances on the FP reduction application using classification between masses and normal tissues in mammography CAD systems.
Similar content being viewed by others
References
Kopans DB. Breast imaging. 3rd ed. Philadelphia: Lippincott Williams & Wilkins; 2007.
Kwon HJ, Shin BH, Gopaul D, Fienberg S. Strain ratio vs. modulus ratio for the diagnosis of breast cancer using elastography. Biomed Eng Lett. 2014; 4(3):292–300.
Mert A, Kilic N, Akan A. An improved hybrid feature reduction for increased breast cancer diagnostic performance. Biomed Eng Lett. 2014; 4(3):285–91.
Suri JS, Rangayyan RM. Recent advances in breast imaging, mammography, and computer-aided diagnosis of breast cancer. Washington: SPIE PRESS; 2006.
Sampat MP. Computer-aided detection and diagnosis in mammography. In: Bovik AC. Handbook of image and video processing. 2nd ed. New York: Academic; 2005. pp. 1195–217.
Tang J, Rangayyan RM, Xu J, El Naqa I, Yang Y. Computer-aided detection and diagnosis of breast cancer with mammography: recent advances. IEEE Trans Inf Technol Biomed. 2009; 13(2):236–51.
Chan HP, Sahiner B, Wagner RF, Petrick N. Classifier design for computer-aided diagnosis: effects of finite sample size on the mean performance of classical and neural network classifiers. Med Phys. 1999; 26(12):2654–88.
Cheng HD, Shi XJ, Min R, Hu LM, Cai XP, Du HN. Approaches for automated detection and classification of masses in mammograms. Pattern Recognit. 2006; 39(4):646–68.
Berber T, Alpkocak A, Balci P, Dicle O. Breast mass contour segmentation algorithm in digital mammograms. Comput Methods Programs Biomed. 2013; 110(2):150–9.
Bilska-Wolak AO, Floyd CE Jr. Tolerance to missing data using a likelihood ratio based classifier for computer-aided classification of breast cancer. Phys Med Biol. 2004; 49(18):4219–37.
Mudigonda NR, Rangayyan RM, Desautels JEL. Gradient and texture analysis for the classification of mammographic masses. IEEE Trans Med Imaging. 2000; 19(10):1032–43.
Jesneck JL, Nolte LW, Baker JA, Floyd CE, Lo JY. Optimized approach to decision fusion of heterogeneous data for breast cancer diagnosis. Med Phys. 2006; 33(8):2945–54.
Wei D, Chan HP, Petrick N, Sahiner B, Helvie MA, Adler DD, Goodsitt MM. False-positive reduction technique for detection of masses on digital mammograms: global and local multiresolution texture analysis. Med Phys. 1997; 24(6):903–14.
Kuncheva LI. Combining pattern classifiers: methods and algorithms. New York: Wiley; 2004.
Freund Y, Schapire RE. A decision-theoretic generalization of on-line learning and an application to boosting. J Comput Syst Sci. 1997; 55(1):119–39.
Constantinidis AS, Fairhurst MC, Rahman AFR. A new multiexpert decision combination algorithms and its application to the detection of circumscribed masses in digital mammograms. Pattern Recognit. 2001; 34(8):1527–37.
Yoon SJ, Kim SJ. AdaBoost-based multiple SVM-RFE for classification of mammograms in DDSM. IEEE Int Conf Bioinf Biomed Workshops. 2008; 75–82.
Breiman L. Random forests. Mach Learning. 2001; 45(1):5–32.
Lu J, Plataniotis KN, Venetsanopoulos AN, Li SZ. Ensembledbased discriminant learning with boosting for face recognition. IEEE Trans Neural Netw. 2006; 17(1):166–78.
Murua A. Upper bounds for error rates of linear combinations of classifiers. IEEE Trans Pattern Anal Mach Intell. 2002; 24(5):591–602.
Way TW, Sahiner B, Hadjiiski LM, Chan HP. Effect of finite sample size on feature selection and classification: a simulation study. Med Phys. 2010; 37(2):907–20.
Wei L, Yang Y, Nishikawa RM, Jiang Y. A study on several machine-learning methods for classification of malignant and benign clustered microcalcifications. IEEE Trans Med Imaging. 2005; 24(3):371–80.
Sahiner B, Chan HP, Petrick N, Wei D, Helvie MA, Adler DD, Goodsitt MM. Classification of mass and normal breast tissue: a convolution neural network classifier with spatial domain and texture images. IEEE Trans Med Imaging. 1996; 15(5):598–609.
Wei D, Chan HP, Helvie MA, Sahiner B, Petrick N, Adler DD, Goodsitt MM. Classification of mass and normal breast tissue on digital mammograms: multiresolution texture analysis. Med Phys. 1995; 22(9):1501–13.
Mudigonda NR, Rangayyan RM, Desautels JEL. Detection of breast masses in mammograms by density slicing and texture flow-field analysis. IEEE Trans Med Imaging. 2001; 20(12):1215–27.
Kupinski MA, Giger ML. Investigation of regularized neural networks for the computerized detection of mass lesions in digital mammograms. IEEE Int Conf Eng Med Biol Soc (EMBS). 1997; 3:1336–9.
Santo MD, Molinara M, Tortorella F, Vento M. Automatic classification of clustered microcalcifications by a multiple expert system. Pattern Recognit. 2003; 36(7):1467–77.
Oliver A, Torrent A, Llado X, Tortajada M, Tortajada L, Sentis M, Freixenet J, Zwiggelaar R. Automatic microcalcification and cluster detection for digital and digitised mammograms. Knowledge-Based Syst. 2012; 28:68–75.
Arod T, Kurdziel M, Sevre EO, Yuen DA. Pattern recognition techniques for automatic detection of suspicious-looking anomalies in mammograms. Comput Methods Programs Biomed. 2005; 79(2):135–49.
Fung G, Krishnapuram B, Merlet N, Ratner E, Bamberger P, Stoeckel J, Rao RB. Addressing image variability while learning classifiers for detecting clusters of micro-calcifications. Int Conf Digit Mammogr. 2006; 4046:84–91.
Friedman J, Hastie T, Tibshirani R. Additive logistic regression: a statistical view of boosting. Annal Stat. 2000; 28(2):337–407.
Dominguez AR, Nandi AK. Detection of masses in mammograms via statistically based enhancement, multilevel-thresholding segmentation, and region selection. Comput Med Imaging Graph. 2008; 32(4):304–15.
Hong BW, Sohn BS. Segmentation of regions of interest in mammograms in a topographic approach. IEEE Trans Inf Technol Biomed. 2010; 14(1):129–39.
Choi JY, Ro YM. Multiresolution local binary pattern texture analysis combined with variable selection for application to false positive reduction in computer-aided detection of breast masses on mammograms. Phys Med Biol. 2012; 57(21):7029–52.
Sahiner B, Chan HP, Petrick N, Helvie MA, Hadjiiski LM. Improvement of mammographic mass characterization using speculation measures and morphological features. Med Phys. 2001; 28(7):1455–65.
Sahiner B, Chan HP, Petrick N, Helvie MA, Goodsitt MM. Computerized characterization of masses on mammograms: the rubber band straightening transform and texture analysis. Med Phys. 1998; 25(4):516–26.
Shavlik JW, Mooney RJ, Towell GG. Symbolic and neural learning algorithms: an experimental comparison. Mach Learn. 1991; 6(2):111–43.
Raudys SJ, Jain AK. Small sample size effects in statistical pattern recognition: recommendations for practitioners. IEEE Trans Pattern Anal Mach Intell. 1991; 13(3):252–64.
Alimoglu F, Alpaydin E. Combining multiple representations and classifiers for pen-based handwritten digit recognition. Turk J Electr Eng. 2001; 9(1):1–12.
Rokach L. Ensemble-based classifiers. Artif Intell Rev. 2010; 33(1):1–39.
Ranawana R, Palade V. Multi-classifier systems: Review and a roadmap for developers. Int J Hybrid Intell Syst. 2006; 3(1):35–61.
Heath M, Bowyer K, Kopans D, Moore R, Kegelmeyer PJ. The digital database for screening mammography. Int Conf Digit Mammography. 2000; 212–8.
Catarious DM Jr, Baydush AH, Floyd CE Jr. Incorporation of an iterative, linear segmentation routine into a mammographic mass CAD system. Med Phys. 2004; 31(6):1512–20.
Eltonsy NH, Tourassi GD, Elmaghraby AS. A concentric morphology model for the detection of masses in mammography. IEEE Trans Med Imaging. 2007; 26(6):880–9.
Zhou XH, McClish DK, Obuchowski NA. Statistical methods in diagnostic medicine. New York: Wiley-Interscience; 2002.
Sahiner B, Chan HP, Petrick N, Helvie MA, Goodsitt MM. Design of a high-sensitivity classifier based on a genetic algorithm: application to computer-aided diagnosis. Phys Med Biol. 1998; 43(10):2853–71.
Vapnik VN. Statistical learning theory. New York: Wiley; 1998.
Setiono R. Feedforward neural network construction using cross validation. Neural Comput. 2001; 13(12):2865–77.
Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol. 2011; 2(3):1–27.
Jain A, Nandakumar K, Ross A. Score normalization in multimodal biometric systems. Pattern Recognit. 2005; 38(12):2270–85.
Mangai UG, Samanta S, Das S, Chowdhury PR. A survey of decision fusion and feature fusion strategies for pattern classification. IETE Tech Rev. 2010; 27(4):293–307.
Nishikawa RM. Current status and future directions of computeraided diagnosis in mammography. Comput Med Imaging Graph. 2007; 31(4):5–224.
Ruta D, Gabrys B. Classifier selection for majority voting. Inf Fusion. 2005; 6(1):63–81.
Wei X, Zhou C, Zhang Q. ICA-based feature fusoin for face recognition. Int J Innov Comput Inf Control. 2010; 6(10):4651–61.
Naik GR, Kumar DK. An overview of independent component analysis and its applications. Inform. 2011; 35(1):63–81.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Choi, J.Y. A generalized multiple classifier system for improving computer-aided classification of breast masses in mammography. Biomed. Eng. Lett. 5, 251–262 (2015). https://doi.org/10.1007/s13534-015-0191-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13534-015-0191-1