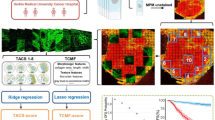
Abstract
Purpose
Collagen features in breast tumor microenvironment is closely associated with the prognosis of patients. We aim to explore the prognostic significance of collagen features at breast tumor border by combining multiphoton imaging and imaging analysis.
Methods
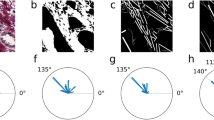
We used multiphoton microscopy (MPM) to label-freely image human breast tumor samples and then constructed an automatic classification model based on deep learning to identify collagen signatures from multiphoton images. We recognized three kinds of collagen signatures at tumor boundary (CSTB I-III) in a small-scale, and furthermore obtained a CSTB score for each patient based on the combined CSTB I-III by using the ridge regression analysis. The prognostic performance of CSTB score is assessed by the area under the receiver operating characteristic curve (AUC), Cox proportional hazard regression analysis, as well as Kaplan-Meier survival analysis.
Results
As an independent prognostic factor, statistical results reveal that the prognostic performance of CSTB score is better than that of the clinical model combining three independent prognostic indicators, molecular subtype, tumor size, and lymph nodal metastasis (AUC, Training dataset: 0.773 vs. 0.749; External validation: 0.753 vs. 0.724; HR, Training dataset: 4.18 vs. 3.92; External validation: 4.98 vs. 4.16), and as an auxiliary indicator, it can greatly improve the accuracy of prognostic prediction. And furthermore, a nomogram combining the CSTB score with the clinical model is established for prognosis prediction and clinical decision making.
Conclusion
This standardized and automated imaging prognosticator may convince pathologists to adopt it as a prognostic factor, thereby customizing more effective treatment plans for patients.




Similar content being viewed by others
Data availability
All data generated or analyzed during this study were included in the article/Supplementary Materials. Further inquiry can be directed to the corresponding authors.
References
H. Korkaya, S. Liu, M.S. Wicha, Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J. Clin. Invest. 121, 3804–3809 (2011). https://doi.org/10.1172/JCI57099
S.D. Soysal, A. Tzankov, S.E. Muenst, Role of the tumor microenvironment in breast cancer. Pathobiology. 82, 142–152 (2015). https://doi.org/10.1159/000430499
B. Ruffell, N.I. Affara, L.M. Coussens, Differential macrophage programming in the tumor microenvironment. Trends Immunol. 33, 119–126 (2012). https://doi.org/10.1016/j.it.2011.12.001
D. Hanahan, L.M. Coussens, Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer cell. 21, 309–322 (2012). https://doi.org/10.1016/j.ccr.2012.02.022
Z. Sun, S.H. Wang, R.C. Zhao, The roles of mesenchymal stem cells in tumor inflammatory microenvironment. J. Hemato Oncol. 7, 1–10 (2014). https://doi.org/10.1186/1756-8722-7-14
S.M. Kakkad, M. Solaiyappan, P. Argani, S. Sukumar, L.K. Jacobs, D. Leibfritz, Z. Bhujwalla, K. Glunde, Collagen I fiber density increases in lymph node positive breast cancers: pilot study. J. Biomed. Opt. 17, 116017 (2012). https://doi.org/10.1117/1.JBO.17.11.116017
C. Frantz, K.M. Stewart, V.M. Weaver, The extracellular matrix at a glance. J. Cell. Sci. 123, 4195–4200 (2010). https://doi.org/10.1242/jcs.023820
G. Charras, E. Sahai, Physical influences of the extracellular environment on cell migration. Nat. Rev. Mol. Cell. Bio. 15, 813–824 (2014). https://doi.org/10.1038/nrm3897
A. Ray, P.P. Provenzano, Aligned forces: Origins and mechanisms of cancer dissemination guided by extracellular matrix architecture. Curr. Opin. Cell. Biol. 72, 63–71 (2021). https://doi.org/10.1016/j.ceb.2021.05.004
L. Gole, J. Yeong, J.C.T. Lim, K.H. Ong, H. Han, A.A. Thike, Y.C. Poh, S. Yee, J. Iqbal, W. Hong, B. Lee, W. Yu, P.H. Tan, Quantitative stain-free imaging and digital profiling of collagen structure reveal diverse survival of triple negative breast cancer patients. Breast Cancer Res. 22, 1–13 (2020). https://doi.org/10.1186/s13058-020-01282-x
R.A. Natal, J. Vassallo, G.R. Paiva, V.B. Pelegati, G.O. Barbosa, G.R. Mendonça, C. Bondarik, S.F. Derchain, H.F. Carvalho, C.S. Lima, C.L. Cesar, L.O. Sarian, Collagen analysis by second-harmonic generation microscopy predicts outcome of luminal breast cancer. Tumor Biology. 40, 1–12 (2018). https://doi.org/10.1177/1010428318770953
M.W. Conklin, J.C. Eickhoff, K.M. Riching, C.A. Pehlke, K.W. Eliceiri, P.P. Provenzano, A. Friedl, P.J. Keely, Aligned collagen is a prognostic signature for survival in human breast carcinoma. AM. J. Pathol. 178, 1221–1232 (2011). https://doi.org/10.1016/j.ajpath.2010.11.076
P.P. Provenzano, K.W. Eliceiri, J.M. Campbell, D.R. Inman, J.G. White, P.J. Keely, Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 4, 1–15 (2016). https://doi.org/10.1186/1741-7015-4-38
M.W. Conklin, R.E. Gangnon, B.L. Sprague, L.V. Gemert, J.M. Hampton, K.W. Eliceiri, J.S. Bredfeldt, Y. Liu, N. Surachaicharn, P.A. Newcomb, A. Friedl, P.J. Keely, Trentham-Dietz, Collagen alignment as a predictor of recurrence after ductal carcinoma in situ. Cancer Epidemiol. Biomarkers Prev. 27, 138–145 (2018). https://doi.org/10.1158/1055-9965.EPI-17-0720
G. Xi, W. Guo, D. Kang, J. Ma, F. Fu, L. Qiu, L. Zheng, J. He, N. Fang, J. Chen, J. Li, S. Zhuo, X. Liao, H. Tu, L. Li, Q. Zhang, C. Wang, S.A. Boppart, J. Chen, Large-scale tumor-associated collagen signatures identify high-risk breast cancer patients. Theranostics. 11, 3229–3243 (2021). https://doi.org/10.7150%2Fthno.55921
L. Li, Z. Han, L. Qiu, D. Kang, Z. Zhan, H. Tu, J. Chen, Evaluation of breast carcinoma regression after preoperative chemotherapy by label-free multiphoton imaging and image analysis. J. Biophotonics. 13, e201900216 (2020). https://doi.org/10.1002/jbio.201900216
B.E. Bejnordi, M. Veta, P.J.V. Diest, B.V. Ginneken, N. Karssemeijer, G. Litjens, A.W.M. Jeroen, Laak, CAMELYON16 Consortium, the, Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama 318, 2199–2210 (2017). https://doi.org/10.1001/jama.2017.14585
B.E. Bejnordi, M. Mullooly, R.M. Pfeiffer, S. Fan, P.M. Vacek, D.L. Weaver, S. Herschorn, L.A. Brinton, B.V. Ginneken, N. Karssemeijer, A.H. Beck, G.L. Gierach, J.A.W.M. Laak, M.E. Sherman, Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod. Pathol. 31, 1502–1512 (2018). https://doi.org/10.1038/s41379-018-0073-z
G. Jiménez, D. Racoceanu, Deep learning for semantic segmentation vs. classification in computational pathology: application to mitosis analysis in breast cancer grading. Front. Bioeng. Biotech. 7, 145 (2019). https://doi.org/10.3389/fbioe.2019.00145
H.D. Couture, L.A. Williams, J. Geradts, S.J. Nyante, E.N. Butler, J.S. Marron, C.M. Perou, M.A. Troester, M. Niethammer, Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. npj Breast Cancer. 4, 30 (2018). https://doi.org/10.1038/s41523-018-0079-1
J.N. Kather, J. Krisam, P. Charoentong, T. Luedde, E. Herpel, C. Weis, T. Gaiser, A. Marx, N.A. Valous, D. Ferber, L. Jansen, C.C. Reyes-Aldasoro, I. Zörnig, D. Jäger, H. Brenner, J. Chang-Claude, M. Hoffmeister, N. Halama, Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. Plos Med. 16 (2019). https://doi.org/10.1371/journal.pmed.1002730. e1002730
N. Naik, A. Madani, A. Esteva, N.S. Keskar, M.F. Press, D. Ruderman, D.B. Agus, R. Socher, Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nat. Commun. 11, 5727 (2020). https://doi.org/10.1038/s41467-020-19334-3
K. He, X. Zhang, S. Ren, J. Sun, Deep residual learning for image recognition. In Proc. IEEE Conference on Computer Vision and Pattern Recognition, 770–778 (2016). https://doi.org/10.48550/arXiv.1512.03385
J. Yang, J. Ju, L. Guo, B. Ji, S. Shi, Z. Yang, S. Gao, X. Yuan, G. Tian, Y. Liang, P. Yuan, Prediction of HER2-positive breast cancer recurrence and metastasis risk from histopathological images and clinical information via multimodal deep learning. Comput. Struct. Biotec. 20, 333–342 (2022). https://doi.org/10.1016/j.csbj.2021.12.028
P. Courtiol, C. Maussion, M. Moarii, E. Pronier, S. Pilcer, M. Sefta, P. Manceron, S. Toldo, M. Zaslavskiy, N.L. Stang, N. Girard, O. Elemento, A.G. Nicholson, J. Blay, F. Galateau-Sallé, G. Wainrib, T. Clozel, Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat. Med. 25, 1519–1525 (2019). https://doi.org/10.1038/s41591-019-0583-3
A. Rasmusso, D. Zilenait, A. Nestarenkaite, R. Augulis, A. Laurinaviciene, V. Ostapenko, T. Poskus, A. Laurinavicius, Immunogradient indicators for antitumor response assessment by automated tumor-stroma interface zone detection. Am. J. Pathol. 190, 1309–1322 (2020). https://doi.org/10.1016/j.ajpath.2020.01.018
E. Terradillos, C.L. Saratxaga, S. Mattana, R. Cicchi, F.S. Pavone, N. Andraka, B.J. Glover, N. Arbide, J. Velasco, M.C. Etxezarraga, A. Picon, Analysis on the characterization of multiphoton microscopy images for malignant neoplastic colon lesion detection under deep learning methods. J. Pathol. Inform. 12, 27 (2021). https://doi.org/10.4103/jpi.jpi_113_20
H. Lin, C. Wei, G. Wang, H. Chen, L. Lin, M. Ni, J. Chen, S. Zhuo, Automated classification of hepatocellular carcinoma differentiation using multiphoton microscopy and deep learning. J. Biophotonics. 12, e201800435 (2019). https://doi.org/10.1002/jbio.201800435
Y. Ogawa, Y. Masugi, T. Abe, K. Yamazaki, A. Ueno, Y. Fujii-Nishimura, S. Hori, H. Yagi, Y. Abe, M. Kitago, M. Sakamoto, Three distinct stroma types in human pancreatic cancer identified by image analysis of fibroblast subpopulations and collagen. Clin. Cancer Res. 27, 107–119 (2021). https://doi.org/10.1158/1078-0432.CCR-20-2298
J. Adur, V.B. Pelegati, A.A. Thomaz, M.O. Baratti, L.A.L.A. Andrade, H.F. Carvalho, F. Bottcher-Luiz, C.L. Cesar, Second harmonic generation microscopy as a powerful diagnostic imaging modality for human ovarian cancer. J. Biophotonics. 7, 37–48 (2014). https://doi.org/10.1002/jbio.201200108
C. He, H. He, J. Chang, B. Chen, H. Ma, M.J. Booth, Polarisation optics for biomedical and clinical applications: a review. Light Sci. Appl. 10, 194 (2021). https://doi.org/10.1038/s41377-021-00639-x
B. Yang, J. Lesicko, M. Sharma, M. Hill, M.S. Sacks, J.W. Tunnell, Polarized light spatial frequency domain imaging for nondestructive quantification of soft tissue fibrous structures. Biomed. Opt. Express. 6, 1520–1533 (2015). https://doi.org/10.1364/BOE.6.001520
S. Manhas, J. Vizet, S. Deby, J.C. Vanel, P. Boito, M. Verdier, A.D. Martino, D. Pagnoux, Demonstration of full 4×4 Mueller polarimetry through an optical fiber for endoscopic applications. Opt. Express. 23, 3047–3054 (2015). https://doi.org/10.1364/OE.23.003047
V. Backman, R. Gurjar, K. Badizadegan, I. Itzkan, R.R. Dasari, L.T. Perelman, M.S. Feld, Polarized light scattering spectroscopy for quantitative measurement of epithelial cellular structures in situ. IEEE J. Sel. Top. Quantum Electron. 5, 1019–1026 (1999). https://doi.org/10.1109/2944.796325
J. Jagtap, S. Chandel, N. Das, J. Soni, S. Chatterjee, A. Pradhan, N. Ghosh, Quantitative Mueller matrix fluorescence spectroscopy for precancer detection. Opt. Lett. 39, 243–246 (2014). https://doi.org/10.1364/OL.39.000243
Acknowledgements
We would like to thank all members of Chen lab for their suggestions and critical feedback.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 82171991. 82172800), Natural Science Foundation of Fujian Province (Nos. 2023J01082, 2023J011125, 2020J01839), Joint Funds for the Innovation of Science and Technology of Fujian Province (2019Y9101).
Author information
Authors and Affiliations
Contributions
Lianhuang Li, Lida Qiu and Jianxin Chen conceived the idea and supervised the study. Xingxin Huang, Xiahui Han, Liqin Zheng, Zhenlin Zhan, Shu Wang and Shunwu Xu performed multiphoton imaging. Fangmeng Fu, Wenhui Guo, Deyong Kang, Qingyuan Zhang, Jianli Ma and Chuan Wang were responsible for sample collection and preparation. Xingxin Huang, Lianhuang Li and Jianxin Chen conducted data analysis, and wrote or revised the manuscript. All the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board at each center in China (Fujian Medical University Union Hospital and Harbin Medical University Cancer Hospital).
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xingxin Huang, Fangmeng Fu and Wenhui Guo contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, X., Fu, F., Guo, W. et al. Prognostic significance of collagen signatures at breast tumor boundary obtained by combining multiphoton imaging and imaging analysis. Cell Oncol. 47, 69–80 (2024). https://doi.org/10.1007/s13402-023-00851-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-023-00851-4