Abstract
Background
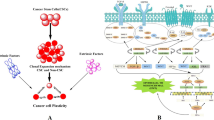
Human malignancies are composed of heterogeneous subpopulations of cancer cells with phenotypic and functional diversity. Among them, a unique subset of cancer stem cells (CSCs) has both the capacity for self-renewal and the potential to differentiate and contribute to multiple tumor properties. As such, CSCs are promising cellular targets for effective cancer therapy. At the molecular level, hyper-activation of multiple stemness regulatory signaling pathways and downstream transcription factors play critical roles in controlling CSCs establishment and maintenance. To regulate CSC properties, these stemness pathways are controlled by post-translational modifications including, but not limited to phosphorylation, acetylation, methylation, and ubiquitination.
Conclusion
In this review, we focus on E3 ubiquitin ligases and their roles and mechanisms in regulating essential hallmarks of CSCs, such as self-renewal, invasion and metastasis, metabolic reprogramming, immune evasion, and therapeutic resistance. Moreover, we discuss emerging therapeutic approaches to eliminate CSCs through targeting E3 ubiquitin ligases by chemical inhibitors and proteolysis-targeting chimera (PROTACs) which are currently under development at the discovery, preclinical, and clinical stages. Several outstanding issues such as roles for E3 ubiquitin ligases in heterogeneity and phenotypical/functional evolution of CSCs remain to be studied under pathologically and clinically relevant conditions. With the rapid application of functional genomic and proteomic approaches at single cell, spatiotemporal, and even single molecule levels, we anticipate that more specific and precise functions of E3 ubiquitin ligases will be delineated in dictating CSC properties. Rational design and proper translation of these mechanistic understandings may lead to novel therapeutic modalities for cancer procession medicine.






Similar content being viewed by others
Data availability
Not applicable.
References
E. Batlle, H. Clevers, Cancer stem cells revisited. Nat. Med. 23(10), 1124–1134 (2017). https://doi.org/10.1038/nm.4409
D. Hanahan, Hallmarks of Cancer: New dimensions. Cancer Discov. 12(1), 31–46 (2022). https://doi.org/10.1158/2159-8290.CD-21-1059
Z.J. Lei, J. Wang, H.L. Xiao, Y. Guo, T. Wang, Q. Li, L. Liu, X. Luo, L.L. Fan, L. Lin, C.Y. Mao, S.N. Wang, Y.L. Wei, C.H. Lan, J. Jiang, X.J. Yang, P.D. Liu, D.F. Chen, B. Wang, Lysine-specific demethylase 1 promotes the stemness and chemoresistance of Lgr5(+) liver cancer initiating cells by suppressing negative regulators of β-catenin signaling. Oncogene 34(24), 3188–3198 (2015). https://doi.org/10.1038/onc.2015.129
Z. Wang, B. Wang, Y. Shi, C. Xu, H.L. Xiao, L.N. Ma, S.L. Xu, L. Yang, Q.L. Wang, W.Q. Dang, W. Cui, S.C. Yu, Y.F. Ping, Y.H. Cui, H.F. Kung, C. Qian, X. Zhang, X.W. Bian, Oncogenic miR-20a and miR-106a enhance the invasiveness of human glioma stem cells by directly targeting TIMP-2. Oncogene 34(11), 1407–1419 (2015). https://doi.org/10.1038/onc.2014.75
L. Deng, T. Meng, L. Chen, W. Wei, P. Wang, The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal 5(1), 11 (2020). https://doi.org/10.1038/s41392-020-0107-0
D. Komander, M. Rape, The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012). https://doi.org/10.1146/annurev-biochem-060310-170328
T.E.T. Mevissen, D. Komander, Mechanisms of Deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192 (2017). https://doi.org/10.1146/annurev-biochem-061516-044916
T. Wang, H. Wu, S. Liu, Z. Lei, Z. Qin, L. Wen, K. Liu, X. Wang, Y. Guo, Q. Liu, L. Liu, J. Wang, L. Lin, C. Mao, X. Zhu, H. Xiao, X. Bian, D. Chen, C. Xu, B. Wang, SMYD3 controls a Wnt-responsive epigenetic switch for ASCL2 activation and cancer stem cell maintenance. Cancer Lett. 430, 11–24 (2018). https://doi.org/10.1016/j.canlet.2018.05.003
T. Wang, Z.Y. Qin, L.Z. Wen, Y. Guo, Q. Liu, Z.J. Lei, W. Pan, K.J. Liu, X.W. Wang, S.J. Lai, W.J. Sun, Y.L. Wei, L. Liu, L. Guo, Y.Q. Chen, J. Wang, H.L. Xiao, X.W. Bian, D.F. Chen, B. Wang, Epigenetic restriction of hippo signaling by MORC2 underlies stemness of hepatocellular carcinoma cells. Cell Death Differ. 25(12), 2086–2100 (2018). https://doi.org/10.1038/s41418-018-0095-6
L. Yang, P. Shi, G. Zhao, J. Xu, W. Peng, J. Zhang, G. Zhang, X. Wang, Z. Dong, F. Chen, H. Cui, Targeting cancer stem cell pathways for cancer therapy. Sig. Transduct. Target. Ther. 5(1), 8 (2020). https://doi.org/10.1038/s41392-020-0110-5
J.A. Clara, C. Monge, Y. Yang, N. Takebe, Targeting signalling pathways and the immune microenvironment of cancer stem cells - a clinical update. Nat. Rev. Clin. Oncol. 17(4), 204–232 (2020). https://doi.org/10.1038/s41571-019-0293-2
B. Wang, Z. Jie, D. Joo, A. Ordureau, P. Liu, W. Gan, J. Guo, J. Zhang, B.J. North, X. Dai, X. Cheng, X. Bian, L. Zhang, J.W. Harper, S.C. Sun, W. Wei, TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature 545(7654), 365–369 (2017). https://doi.org/10.1038/nature22344
A. Strikoudis, M. Guillamot, I. Aifantis, Regulation of stem cell function by protein ubiquitylation. EMBO Rep. 15(4), 365–382 (2014). https://doi.org/10.1002/embr.201338373
N. Cai, M. Li, J. Qu, G.H. Liu, J.C. Izpisua Belmonte, Post-translational modulation of pluripotency. J. Mol. Cell Biol. 4(4), 262–265 (2012). https://doi.org/10.1093/jmcb/mjs031
Y. Kong, Z. Wang, M. Huang, Z. Zhou, Y. Li, H. Miao, X. Wan, J. Huang, X. Mao, C. Chen, CUL7 promotes cancer cell survival through promoting Caspase-8 ubiquitination. Int. J. Cancer 145(5), 1371–1381 (2019). https://doi.org/10.1002/ijc.32239
L. Song, Z.Q. Luo, Post-translational regulation of ubiquitin signaling. J. Cell Biol. 218(6), 1776–1786 (2019). https://doi.org/10.1083/jcb.201902074
S. Haq, B. Suresh, S. Ramakrishna, Deubiquitylating enzymes as cancer stem cell therapeutics. Biochimica et biophysica acta Rev. Cancer 1869(1), 1–10 (2018). https://doi.org/10.1016/j.bbcan.2017.10.004
S.A. Abdul Rehman, Y.A. Kristariyanto, S.Y. Choi, P.J. Nkosi, S. Weidlich, K. Labib, K. Hofmann, Y. Kulathu, MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell 63(1), 146–155 (2016). https://doi.org/10.1016/j.molcel.2016.05.009
M.A. Basar, D.B. Beck, A. Werner, Deubiquitylases in developmental ubiquitin signaling and congenital diseases. Cell Death Differ. 28(2), 538–556 (2021). https://doi.org/10.1038/s41418-020-00697-5
F.E. Morreale, H. Walden, Types of ubiquitin ligases. Cell 165(1), 248–248.e1 (2016). https://doi.org/10.1016/j.cell.2016.03.003
N. Zheng, N. Shabek, Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 (2017). https://doi.org/10.1146/annurev-biochem-060815-014922
D. Rotin, S. Kumar, Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10(6), 398–409 (2009). https://doi.org/10.1038/nrm2690
S. Singh, J. Ng, J. Sivaraman, Exploring the “Other” subfamily of HECT E3-ligases for therapeutic intervention. Pharmacol. Ther. 224, (2021). https://doi.org/10.1016/j.pharmthera.2021.107809
M.Y. Ryu, S.K. Cho, Y. Hong, J. Kim, J.H. Kim, G.M. Kim, Y.J. Chen, E. Knoch, B.L. Møller, W.T. Kim, M.F. Lyngkjær, S.W. Yang, Classification of barley U-box E3 ligases and their expression patterns in response to drought and pathogen stresses. BMC Genomics 20(1), 326 (2019). https://doi.org/10.1186/s12864-019-5696-z
K.K. Dove, R.E. Klevit, RING-between-RING E3 ligases: Emerging themes amid the variations. J. Mol. Biol. 429(22), 3363–3375 (2017). https://doi.org/10.1016/j.jmb.2017.08.008
P. Wang, X. Dai, W. Jiang, Y. Li, W. Wei, RBR E3 ubiquitin ligases in tumorigenesis. Semin. Cancer Biol. 67(Pt 2), 131–144 (2020). https://doi.org/10.1016/j.semcancer.2020.05.002
J. Low, W. Blosser, M. Dowless, L. Ricci-Vitiani, R. Pallini, R. de Maria, L. Stancato, Knockdown of ubiquitin ligases in glioblastoma cancer stem cells leads to cell death and differentiation. J. Biomol. Screen. 17(2), 152–162 (2012). https://doi.org/10.1177/1087057111422565
M. Quiroga, A. Rodríguez-Alonso, G. Alfonsín, J.J.E. Rodríguez, S.M. Breijo, V. Chantada, A. Figueroa, Protein degradation by E3 ubiquitin ligases in Cancer stem cells. Cancers 14(4), 990 (2022). https://doi.org/10.3390/cancers14040990
D. Senft, J. Qi, Z.A. Ronai, Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer 18(2), 69–88 (2018). https://doi.org/10.1038/nrc.2017.105
S. Hume, C.P. Grou, P. Lascaux, V. D'Angiolella, A.J. Legrand, K. Ramadan, G.L. Dianov, The NUCKS1-SKP2-p21/p27 axis controls S phase entry. Nat. Commun. 12(1), 6959 (2021). https://doi.org/10.1038/s41467-021-27124-8
A. Nowosad, P. Jeannot, C. Callot, J. Creff, R.T. Perchey, C. Joffre, P. Codogno, S. Manenti, A. Besson, p27 controls Ragulator and mTOR activity in amino acid-deprived cells to regulate the autophagy-lysosomal pathway and coordinate cell cycle and cell growth. Nat. Cell Biol. 22(9), 1076–1090 (2020). https://doi.org/10.1038/s41556-020-0554-4
J. Deng, X. Bai, X. Feng, J. Ni, J. Beretov, P. Graham, Y. Li, Inhibition of PI3K/Akt/mTOR signaling pathway alleviates ovarian cancer chemoresistance through reversing epithelial-mesenchymal transition and decreasing cancer stem cell marker expression. BMC Cancer 19(1), 618 (2019). https://doi.org/10.1186/s12885-019-5824-9
C.H. Chan, C.F. Li, W.L. Yang, Y. Gao, S.W. Lee, Z. Feng, H.Y. Huang, K.K.C. Tsai, L.G. Flores, Y. Shao, J.D. Hazle, D. Yu, W. Wei, D. Sarbassov, M.C. Hung, K.I. Nakayama, H.K. Lin, The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, Herceptin sensitivity, and tumorigenesis. Cell 151(4), 913–914 (2012). https://doi.org/10.1016/j.cell.2012.10.025
W.L. Yang, J. Wang, C.H. Chan, S.W. Lee, A.D. Campos, B. Lamothe, L. Hur, B.C. Grabiner, X. Lin, B.G. Darnay, H.K. Lin, The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science (New York, N.Y.) 325(5944), 1134–1138 (2009). https://doi.org/10.1126/science.1175065
C.H. Chan, J.K. Morrow, C.F. Li, Y. Gao, G. Jin, A. Moten, L.J. Stagg, J.E. Ladbury, Z. Cai, D. Xu, C.J. Logothetis, M.C. Hung, S. Zhang, H.K. Lin, Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell 154(3), 556–568 (2013). https://doi.org/10.1016/j.cell.2013.06.048
C.H. Yeh, M. Bellon, C. Nicot, FBXW7: A critical tumor suppressor of human cancers. Mol. Cancer 17(1), 115 (2018). https://doi.org/10.1186/s12943-018-0857-2
L. Reavie, S.M. Buckley, E. Loizou, S. Takeishi, B. Aranda-Orgilles, D. Ndiaye-Lobry, O. Abdel-Wahab, S. Ibrahim, K.I. Nakayama, I. Aifantis, Regulation of c-Myc ubiquitination controls chronic myelogenous leukemia initiation and progression. Cancer Cell 23(3), 362–375 (2013). https://doi.org/10.1016/j.ccr.2013.01.025
Y. Zhao, X. Xiong, Y. Sun, Cullin-RING ligase 5: Functional characterization and its role in human cancers. Semin. Cancer Biol. 67(Pt 2), 61–79 (2020). https://doi.org/10.1016/j.semcancer.2020.04.003
X. Hong, H.T. Nguyen, Q. Chen, R. Zhang, Z. Hagman, P.M. Voorhoeve, S.M. Cohen, Opposing activities of the Ras and Hippo pathways converge on regulation of YAP protein turnover. EMBO J. 33(21), 2447–2457 (2014). https://doi.org/10.15252/embj.201489385
B. Cui, L. Gong, M. Chen, Y. Zhang, H. Yuan, J. Qin, D. Gao, CUL5-SOCS6 complex regulates mTORC2 function by targeting Sin1 for degradation. Cell Discov. 5, 52 (2019). https://doi.org/10.1038/s41421-019-0118-6
T. Yoshizumi, A. Kubo, H. Murata, M. Shinonaga, H. Kanno, BC-box motif in SOCS6 induces differentiation of epidermal stem cells into GABAnergic neurons. Int. J. Mol. Sci. 21(14), 4947 (2020). https://doi.org/10.3390/ijms21144947
C. Dominguez-Brauer, R. Khatun, A.J. Elia, K.L. Thu, P. Ramachandran, S.P. Baniasadi, Z. Hao, L.D. Jones, J. Haight, Y. Sheng, T.W. Mak, E3 ubiquitin ligase Mule targets β-catenin under conditions of hyperactive Wnt signaling. Proc. Natl. Acad. Sci. U. S. A. 114(7), E1148–E1157 (2017). https://doi.org/10.1073/pnas.1621355114
K.B. Myant, P. Cammareri, M.C. Hodder, J. Wills, A. Von Kriegsheim, B. Győrffy, M. Rashid, S. Polo, E. Maspero, L. Vaughan, B. Gurung, E. Barry, A. Malliri, F. Camargo, D.J. Adams, A. Iavarone, A. Lasorella, O.J. Sansom, HUWE1 is a critical colonic tumour suppressor gene that prevents MYC signalling, DNA damage accumulation and tumour initiation. EMBO Mol. Med. 9(2), 181–197 (2017). https://doi.org/10.15252/emmm.201606684
J. Zhang, M. Chen, Y. Zhu, X. Dai, F. Dang, J. Ren, S. Ren, Y.V. Shulga, F. Beca, W. Gan, F. Wu, Y.M. Lin, X. Zhou, J.A. DeCaprio, A.H. Beck, K.P. Lu, J. Huang, C. Zhao, Y. Sun, X. Gao, … W. Wei, SPOP promotes Nanog destruction to suppress stem cell traits and prostate Cancer progression. Dev. Cell. 48(3), 329–344.e5 (2019). https://doi.org/10.1016/j.devcel.2018.11.035
X. Wang, J. Jin, F. Wan, L. Zhao, H. Chu, C. Chen, G. Liao, J. Liu, Y. Yu, H. Teng, L. Fang, C. Jiang, W. Pan, X. Xie, J. Li, X. Lu, X. Jiang, X. Ge, D. Ye, P. Wang, AMPK promotes SPOP-mediated NANOG degradation to regulate prostate Cancer cell Stemness. Dev. Cell 48(3), 345–360.e7 (2019). https://doi.org/10.1016/j.devcel.2018.11.033
P. Tan, Y. Xu, Y. Du, L. Wu, B. Guo, S. Huang, J. Zhu, B. Li, F. Lin, L. Yao, SPOP suppresses pancreatic cancer progression by promoting the degradation of NANOG. Cell Death Dis. 10(11), 794 (2019). https://doi.org/10.1038/s41419-019-2017-z
X. Dai, W. Gan, X. Li, S. Wang, W. Zhang, L. Huang, S. Liu, Q. Zhong, J. Guo, J. Zhang, T. Chen, K. Shimizu, F. Beca, M. Blattner, D. Vasudevan, D.L. Buckley, J. Qi, L. Buser, P. Liu, H. Inuzuka, … W. Wei, Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of Na. Nat. Med. 23(9), 1063–1071 (2017). https://doi.org/10.1038/nm.4378
X. Dai, Z. Wang, W. Wei, SPOP-mediated degradation of BRD4 dictates cellular sensitivity to BET inhibitors. Cell Cycle (Georgetown, Tex) 16(24), 2326–2329 (2017). https://doi.org/10.1080/15384101.2017.1388973
P. Zhang, D. Wang, Y. Zhao, S. Ren, K. Gao, Z. Ye, S. Wang, C.W. Pan, Y. Zhu, Y. Yan, Y. Yang, D. Wu, Y. He, J. Zhang, D. Lu, X. Liu, L. Yu, S. Zhao, Y. Li, D. Lin, … H. Huang, Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nat. Med. 23(9), 1055–1062 (2017). https://doi.org/10.1038/nm.4379
L.G. Ju, Y. Zhu, Q.Y. Long, X.J. Li, X. Lin, S.B. Tang, L. Yin, Y. Xiao, X.H. Wang, L. Li, L. Zhang, M. Wu, SPOP suppresses prostate cancer through regulation of CYCLIN E1 stability. Cell Death Differ. 26(6), 1156–1168 (2019). https://doi.org/10.1038/s41418-018-0198-0
O.A. Guryanova, Q. Wu, L. Cheng, J.D. Lathia, Z. Huang, J. Yang, J. MacSwords, C.E. Eyler, R.E. McLendon, J.M. Heddleston, W. Shou, D. Hambardzumyan, J. Lee, A.B. Hjelmeland, A.E. Sloan, M. Bredel, G.R. Stark, J.N. Rich, S. Bao, Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell 19(4), 498–511 (2011). https://doi.org/10.1016/j.ccr.2011.03.004
C. Zhang, S. Mukherjee, C. Tucker-Burden, J.L. Ross, M.J. Chau, J. Kong, D.J. Brat, TRIM8 regulates stemness in glioblastoma through PIAS3-STAT3. Mol. Oncol. 11(3), 280–294 (2017). https://doi.org/10.1002/1878-0261.12034
J. Yao, T. Xu, T. Tian, X. Fu, W. Wang, S. Li, T. Shi, A. Suo, Z. Ruan, H. Guo, Y. Yao, Tripartite motif 16 suppresses breast cancer stem cell properties through regulation of Gli-1 degradation via the ubiquitin-proteasome pathway. Oncol. Rep. 35(2), 1204–1212 (2016). https://doi.org/10.3892/or.2015.4437
P. Czerwińska, P.K. Shah, K. Tomczak, M. Klimczak, S. Mazurek, B. Sozańska, P. Biecek, K. Korski, V. Filas, A. Mackiewicz, J.N. Andersen, M. Wiznerowicz, TRIM28 multi-domain protein regulates cancer stem cell population in breast tumor development. Oncotarget 8(1), 863–882 (2017). https://doi.org/10.18632/oncotarget.13273
P. Czerwińska, S. Mazurek, M. Wiznerowicz, The complexity of TRIM28 contribution to cancer. J. Biomed. Sci. 24(1), 63 (2017). https://doi.org/10.1186/s12929-017-0374-4
S. Wiszniak, S. Kabbara, R. Lumb, M. Scherer, G. Secker, N. Harvey, S. Kumar, Q. Schwarz, The ubiquitin ligase Nedd4 regulates craniofacial development by promoting cranial neural crest cell survival and stem-cell like properties. Dev. Biol. 383(2), 186–200 (2013). https://doi.org/10.1016/j.ydbio.2013.09.024
L.C. Trotman, X. Wang, A. Alimonti, Z. Chen, J. Teruya-Feldstein, H. Yang, N.P. Pavletich, B.S. Carver, C. Cordon-Cardo, H. Erdjument-Bromage, P. Tempst, S.G. Chi, H.J. Kim, T. Misteli, X. Jiang, P.P. Pandolfi, Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128(1), 141–156 (2007). https://doi.org/10.1016/j.cell.2006.11.040
M. Yue, Z. Yun, S. Li, G. Yan, Z. Kang, NEDD4 triggers FOXA1 ubiquitination and promotes colon cancer progression under microRNA-340-5p suppression and ATF1 upregulation. RNA Biol. 18(11), 1981–1995 (2021). https://doi.org/10.1080/15476286.2021.1885232
C. Lu, C. Thoeni, A. Connor, H. Kawabe, S. Gallinger, D. Rotin, Intestinal knockout of Nedd4 enhances growth of Apcmin tumors. Oncogene 35(45), 5839–5849 (2016). https://doi.org/10.1038/onc.2016.125
L. Novellasdemunt, A. Kucharska, C. Jamieson, M. Prange-Barczynska, A. Baulies, P. Antas, J. van der Vaart, H. Gehart, M.M. Maurice, V.S. Li, NEDD4 and NEDD4L regulate Wnt signalling and intestinal stem cell priming by degrading LGR5 receptor. EMBO J. 39(3), e102771 (2020). https://doi.org/10.15252/embj.2019102771
J. Chen, A. Mitra, S. Li, S. Song, B.N. Nguyen, J.S. Chen, J.H. Shin, N.R. Gough, P. Lin, V. Obias, A.R. He, Z. Yao, T.M. Malta, H. Noushmehr, P.S. Latham, X. Su, A. Rashid, B. Mishra, R.C. Wu, L. Mishra, Targeting the E3 ubiquitin ligase PJA1 enhances tumor-suppressing TGFβ signaling. Cancer Res. 80(9), 1819–1832 (2020). https://doi.org/10.1158/0008-5472.CAN-19-3116
J. Chen, Z.X. Yao, J.S. Chen, Y.J. Gi, N.M. Muñoz, S. Kundra, H.F. Herlong, Y.S. Jeong, A. Goltsov, K. Ohshiro, N.A. Mistry, J. Zhang, X. Su, S. Choufani, A. Mitra, S. Li, B. Mishra, J. White, A. Rashid, A.Y. Wang, … L. Mishra, TGF-β/β2-spectrin/CTCF-regulated tumor suppression in human stem cell disorder Beckwith-Wiedemann syndrome. J. Clin. Investig. 126(2), 527–542 (2016). https://doi.org/10.1172/JCI80937
J. Gu, W. Mao, W. Ren, F. Xu, Q. Zhu, C. Lu, Z. Lin, Z. Zhang, Y. Chu, R. Liu, D. Ge, Ubiquitin-protein ligase E3C maintains non-small-cell lung cancer stemness by targeting AHNAK-p53 complex. Cancer Lett. 443, 125–134 (2019). https://doi.org/10.1016/j.canlet.2018.11.029
F. Yang, Y. Xing, Y. Li, X. Chen, J. Jiang, Z. Ai, Y. Wei, Monoubiquitination of Cancer stem cell marker CD133 at lysine 848 regulates its secretion and promotes cell migration. Mol. Cell. Biol. 38(15), e00024–e00018 (2018). https://doi.org/10.1128/MCB.00024-18
M. Lin, J. Pan, Q. Chen, Z. Xu, X. Lin, C. Shi, Overexpression of FOXA1 inhibits cell proliferation and EMT of human gastric cancer AGS cells. Gene 642, 145–151 (2018). https://doi.org/10.1016/j.gene.2017.11.023
H.E. Hsia, R. Kumar, R. Luca, M. Takeda, J. Courchet, J. Nakashima, S. Wu, S. Goebbels, W. An, B.J. Eickholt, F. Polleux, D. Rotin, H. Wu, M.J. Rossner, C. Bagni, J.S. Rhee, N. Brose, H. Kawabe, Ubiquitin E3 ligase Nedd4-1 acts as a downstream target of PI3K/PTEN-mTORC1 signaling to promote neurite growth. Proc. Natl. Acad. Sci. U. S. A. 111(36), 13205–13210 (2014). https://doi.org/10.1073/pnas.1400737111
A. Porčnik, M. Novak, B. Breznik, B. Majc, B. Hrastar, N. Šamec, A. Zottel, I. Jovčevska, M. Vittori, A. Rotter, R. Komel, T. Lah Turnšek, TRIM28 selective Nanobody reduces glioblastoma stem cell invasion. Molecules (Basel, Switzerland) 26(17), 5141 (2021). https://doi.org/10.3390/molecules26175141
Y. Zhang, X. Zhang, M. Ye, P. Jing, J. Xiong, Z. Han, J. Kong, M. Li, X. Lai, N. Chang, J. Zhang, J. Zhang, FBW7 loss promotes epithelial-to-mesenchymal transition in non-small cell lung cancer through the stabilization of Snail protein. Cancer Lett. 419, 75–83 (2018). https://doi.org/10.1016/j.canlet.2018.01.047
Y. Wang, Y. Liu, J. Lu, P. Zhang, Y. Wang, Y. Xu, Z. Wang, J.H. Mao, G. Wei, Rapamycin inhibits FBXW7 loss-induced epithelial-mesenchymal transition and cancer stem cell-like characteristics in colorectal cancer cells. Biochem. Biophys. Res. Commun. 434(2), 352–356 (2013). https://doi.org/10.1016/j.bbrc.2013.03.077
P. Gulhati, K.A. Bowen, J. Liu, P.D. Stevens, P.G. Rychahou, M. Chen, E.Y. Lee, H.L. Weiss, K.L. O'Connor, T. Gao, B.M. Evers, mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 71(9), 3246–3256 (2011). https://doi.org/10.1158/0008-5472.CAN-10-4058
A.B. D'Assoro, T. Liu, C. Quatraro, A. Amato, M. Opyrchal, A. Leontovich, Y. Ikeda, S. Ohmine, W. Lingle, V. Suman, J. Ecsedy, I. Iankov, A. Di Leonardo, J. Ayers-Inglers, A. Degnim, D. Billadeau, J. McCubrey, J. Ingle, J.L. Salisbury, E. Galanis, The mitotic kinase Aurora--a promotes distant metastases by inducing epithelial-to-mesenchymal transition in ERα(+) breast cancer cells. Oncogene 33(5), 599–610 (2014). https://doi.org/10.1038/onc.2012.628
J.H. Wald, J. Hatakeyama, I. Printsev, A. Cuevas, W.H.D. Fry, M.J. Saldana, K. VanderVorst, A. Rowson-Hodel, J.M. Angelastro, C. Sweeney, K.L. Carraway, Suppression of planar cell polarity signaling and migration in glioblastoma by Nrdp1-mediated Dvl polyubiquitination. Oncogene 36(36), 5158–5167 (2017). https://doi.org/10.1038/onc.2017.126
C.Y. Wei, M.X. Zhu, Y.W. Yang, P.F. Zhang, X. Yang, R. Peng, C. Gao, J.C. Lu, L. Wang, X.Y. Deng, N.H. Lu, F.Z. Qi, J.Y. Gu, Downregulation of RNF128 activates Wnt/β-catenin signaling to induce cellular EMT and stemness via CD44 and CTTN ubiquitination in melanoma. J. Hematol. Oncol. 12(1), 21 (2019). https://doi.org/10.1186/s13045-019-0711-z
Y. Jin, X. Zhao, Q. Zhang, Y. Zhang, X. Fu, X. Hu, Y. Wan, Cancer-associated mutation abolishes the impact of TRIM21 on the invasion of breast cancer cells. Int. J. Biol. Macromol. 142, 782–789 (2020). https://doi.org/10.1016/j.ijbiomac.2019.10.019
C. Jiao, T. Meng, C. Zhou, X. Wang, P. Wang, M. Lu, X. Tan, Q. Wei, X. Ge, J. Jin, TGF-β signaling regulates SPOP expression and promotes prostate cancer cell stemness. Aging 12(9), 7747–7760 (2020). https://doi.org/10.18632/aging.103085
P. Ekambaram, J.L. Lee, N.E. Hubel, D. Hu, S. Yerneni, P.G. Campbell, N. Pollock, L.R. Klei, V.J. Concel, P.C. Delekta, A.M. Chinnaiyan, S.A. Tomlins, D.R. Rhodes, N. Priedigkeit, A.V. Lee, S. Oesterreich, L.M. McAllister-Lucas, P.C. Lucas, The CARMA3-Bcl10-MALT1 signalosome drives NFκB activation and promotes aggressiveness in angiotensin II receptor-positive breast Cancer. Cancer Res. 78(5), 1225–1240 (2018). https://doi.org/10.1158/0008-5472.CAN-17-1089
Y.F. Huang, Z. Zhang, M. Zhang, Y.S. Chen, J. Song, P.F. Hou, H.M. Yong, J.N. Zheng, J. Bai, CUL1 promotes breast cancer metastasis through regulating EZH2-induced the autocrine expression of the cytokines CXCL8 and IL11. Cell Death Dis. 10(1), 2 (2018). https://doi.org/10.1038/s41419-018-1258-6
J. Liu, C. Zhang, Y. Zhao, X. Yue, H. Wu, S. Huang, J. Chen, K. Tomsky, H. Xie, C.A. Khella, M.L. Gatza, D. Xia, J. Gao, E. White, B.G. Haffty, W. Hu, Z. Feng, Parkin targets HIF-1α for ubiquitination and degradation to inhibit breast tumor progression. Nat. Commun. 8(1), 1823 (2017). https://doi.org/10.1038/s41467-017-01947-w
N.N. Pavlova, C.B. Thompson, The emerging hallmarks of Cancer metabolism. Cell Metab. 23(1), 27–47 (2016). https://doi.org/10.1016/j.cmet.2015.12.006
E.J. Kim, S.H. Kim, X. Jin, X. Jin, H. Kim, KCTD2, an adaptor of Cullin3 E3 ubiquitin ligase, suppresses gliomagenesis by destabilizing c-Myc. Cell Death Differ. 24(4), 649–659 (2017). https://doi.org/10.1038/cdd.2016.151
V.B. Pillai, N.R. Sundaresan, M.P. Gupta, Regulation of Akt signaling by sirtuins: Its implication in cardiac hypertrophy and aging. Circ. Res. 114(2), 368–378 (2014). https://doi.org/10.1161/CIRCRESAHA.113.300536
X. Jin, Y. Pan, L. Wang, L. Zhang, R. Ravichandran, P.R. Potts, J. Jiang, H. Wu, H. Huang, MAGE-TRIM28 complex promotes the Warburg effect and hepatocellular carcinoma progression by targeting FBP1 for degradation. Oncogenesis 6(4), e312 (2017). https://doi.org/10.1038/oncsis.2017.21
Y. Hou, F. Moreau, K. Chadee, PPARγ is an E3 ligase that induces the degradation of NFκB/p65. Nat. Commun. 3, 1300 (2012). https://doi.org/10.1038/ncomms2270
Z. Zhao, D. Xu, Z. Wang, L. Wang, R. Han, Z. Wang, L. Liao, Y. Chen, Hepatic PPARα function is controlled by polyubiquitination and proteasome-mediated degradation through the coordinated actions of PAQR3 and HUWE1. Hepatology (Baltimore, Md) 68(1), 289–303 (2018). https://doi.org/10.1002/hep.29786
Y. Shang, J. He, Y. Wang, Q. Feng, Y. Zhang, J. Guo, J. Li, S. Li, Y. Wang, G. Yan, F. Ren, Y. Shi, J. Xu, N. Zeps, Y. Zhai, D. He, Z. Chang, CHIP/Stub1 regulates the Warburg effect by promoting degradation of PKM2 in ovarian carcinoma. Oncogene 36(29), 4191–4200 (2017). https://doi.org/10.1038/onc.2017.31
S. Amin, P. Yang, Z. Li, Pyruvate kinase M2: A multifarious enzyme in non-canonical localization to promote cancer progression. Biochimica et biophysica acta. Rev. Cancer 1871(2), 331–341 (2019). https://doi.org/10.1016/j.bbcan.2019.02.003
J. Schödel, S. Grampp, E.R. Maher, H. Moch, P.J. Ratcliffe, P. Russo, D.R. Mole, Hypoxia, hypoxia-inducible transcription factors, and renal Cancer. Eur. Urol. 69(4), 646–657 (2016). https://doi.org/10.1016/j.eururo.2015.08.007
S. Ros, A. Schulze, Glycolysis back in the limelight: Systemic targeting of HK2 blocks tumor growth. Cancer Discov. 3(10), 1105–1107 (2013). https://doi.org/10.1158/2159-8290.CD-13-0565
L. Jiao, H.L. Zhang, D.D. Li, K.L. Yang, J. Tang, X. Li, J. Ji, Y. Yu, R.Y. Wu, S. Ravichandran, J.J. Liu, G.K. Feng, M.S. Chen, Y.X. Zeng, R. Deng, X.F. Zhu, Regulation of glycolytic metabolism by autophagy in liver cancer involves selective autophagic degradation of HK2 (hexokinase 2). Autophagy 14(4), 671–684 (2018). https://doi.org/10.1080/15548627.2017.1381804
S. Yoshino, T. Hara, H.J. Nakaoka, A. Kanamori, Y. Murakami, M. Seiki, T. Sakamoto, The ERK signaling target RNF126 regulates anoikis resistance in cancer cells by changing the mitochondrial metabolic flux. Cell Discov. 2, 16019 (2016). https://doi.org/10.1038/celldisc.2016.19
A.M. Roberts, D.K. Miyamoto, T.R. Huffman, L.A. Bateman, A.N. Ives, D. Akopian, M.J. Heslin, C.M. Contreras, M. Rape, C.F. Skibola, D.K. Nomura, Chemoproteomic screening of covalent ligands reveals UBA5 as a novel pancreatic Cancer target. ACS Chem. Biol. 12(4), 899–904 (2017). https://doi.org/10.1021/acschembio.7b00020
C.J. David, J. Massagué, Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 19(7), 419–435 (2018). https://doi.org/10.1038/s41580-018-0007-0
Y. Yang, M. Luo, K. Zhang, J. Zhang, T. Gao, D.O. Connell, F. Yao, C. Mu, B. Cai, Y. Shang, W. Chen, Nedd4 ubiquitylates VDAC2/3 to suppress erastin-induced ferroptosis in melanoma. Nat. Commun. 11(1), 433 (2020). https://doi.org/10.1038/s41467-020-14324-x
Y. Wang, Y. Liu, J. Liu, R. Kang, D. Tang, NEDD4L-mediated LTF protein degradation limits ferroptosis. Biochem. Biophys. Res. Commun. 531(4), 581–587 (2020). https://doi.org/10.1016/j.bbrc.2020.07.032
Z. Ye, Q. Zhuo, Q. Hu, X. Xu, M. Liu, Z. Zhang, W. Xu, W. Liu, G. Fan, Y. Qin, X. Yu, S. Ji, FBW7-NRA41-SCD1 axis synchronously regulates apoptosis and ferroptosis in pancreatic cancer cells. Redox Biol. 38, 101807 (2021). https://doi.org/10.1016/j.redox.2020.101807
J. Koo, P. Yue, X. Deng, F.R. Khuri, S.Y. Sun, mTOR complex 2 stabilizes Mcl-1 protein by suppressing its glycogen synthase kinase 3-dependent and SCF-FBXW7-mediated degradation. Mol. Cell. Biol. 35(13), 2344–2355 (2015). https://doi.org/10.1128/MCB.01525-14
P. Tsvetkov, S. Coy, B. Petrova, M. Dreishpoon, A. Verma, M. Abdusamad, J. Rossen, L. Joesch-Cohen, R. Humeidi, R.D. Spangler, J.K. Eaton, E. Frenkel, M. Kocak, S.M. Corsello, S. Lutsenko, N. Kanarek, S. Santagata, T.R. Golub, Copper induces cell death by targeting lipoylated TCA cycle proteins. Science (New York, N.Y.) 375(6586), 1254–1261 (2022). https://doi.org/10.1126/science.abf0529
B. Zhang, T. Binks, R. Burke, The E3 ubiquitin ligase Slimb/β-TrCP is required for normal copper homeostasis in Drosophila. Biochimica et biophysica acta. Mol. Cell Res. 1867(10), 118768 (2020). https://doi.org/10.1016/j.bbamcr.2020.118768
B.Z. Carter, P.Y. Mak, H. Mu, X. Wang, W. Tao, D.H. Mak, E.J. Dettman, M. Cardone, O. Zernovak, T. Seki, M. Andreeff, Combined inhibition of MDM2 and BCR-ABL1 tyrosine kinase targets chronic myeloid leukemia stem/progenitor cells in a murine model. Haematologica 105(5), 1274–1284 (2020). https://doi.org/10.3324/haematol.2019.219261
D. Venkatesh, N.A. O'Brien, F. Zandkarimi, D.R. Tong, M.E. Stokes, D.E. Dunn, E.S. Kengmana, A.T. Aron, A.M. Klein, J.M. Csuka, S.H. Moon, M. Conrad, C.J. Chang, D.C. Lo, A. D'Alessandro, C. Prives, B.R. Stockwell, MDM2 and MDMX promote ferroptosis by PPARα-mediated lipid remodeling. Genes Dev. 34(7–8), 526–543 (2020). https://doi.org/10.1101/gad.334219.119
M.S. Schrock, B.R. Stromberg, L. Scarberry, M.K. Summers, APC/C ubiquitin ligase: Functions and mechanisms in tumorigenesis. Semin. Cancer Biol. 67(Pt 2), 80–91 (2020). https://doi.org/10.1016/j.semcancer.2020.03.001
P. Zhang, K. Gao, L. Zhang, H. Sun, X. Zhao, Y. Liu, Z. Lv, Q. Shi, Y. Chen, D. Jiao, Y. Li, W. Gu, C. Wang, CRL2-KLHDC3 E3 ubiquitin ligase complex suppresses ferroptosis through promoting p14ARF degradation. Cell Death Differ. 29(4), 758–771 (2022). https://doi.org/10.1038/s41418-021-00890-0
E. Villa, R. Paul, O. Meynet, S. Volturo, G. Pinna, J.E. Ricci, The E3 ligase UBR2 regulates cell death under caspase deficiency via Erk/MAPK pathway. Cell Death Dis. 11(12), 1041 (2020). https://doi.org/10.1038/s41419-020-03258-3
A. Sato, J. Sunayama, K. Matsuda, S. Seino, K. Suzuki, E. Watanabe, K. Tachibana, A. Tomiyama, T. Kayama, C. Kitanaka, MEK-ERK signaling dictates DNA-repair gene MGMT expression and temozolomide resistance of stem-like glioblastoma cells via the MDM2-p53 axis. Stem Cells (Dayton, Ohio) 29(12), 1942–1951 (2011). https://doi.org/10.1002/stem.753
D. Izumi, T. Ishimoto, K. Miyake, T. Eto, K. Arima, Y. Kiyozumi, T. Uchihara, J. Kurashige, M. Iwatsuki, Y. Baba, Y. Sakamoto, Y. Miyamoto, N. Yoshida, M. Watanabe, A. Goel, P. Tan, H. Baba, Colorectal Cancer stem cells acquire Chemoresistance through the upregulation of F-box/WD repeat-containing protein 7 and the consequent degradation of c-Myc. Stem Cells (Dayton, Ohio) 35(9), 2027–2036 (2017). https://doi.org/10.1002/stem.2668
Z. Zhao, Y. Wang, D. Yun, Q. Huang, D. Meng, Q. Li, P. Zhang, C. Wang, H. Chen, D. Lu, TRIM21 overexpression promotes tumor progression by regulating cell proliferation, cell migration and cell senescence in human glioma. Am. J. Cancer Res. 10(1), 114–130 (2020)
M. Maugeri-Saccà, P. Vigneri, R. De Maria, Cancer stem cells and chemosensitivity. Clin. Cancer Res. 17(15), 4942–4947 (2011). https://doi.org/10.1158/1078-0432.CCR-10-2538
M. Cojoc, K. Mäbert, M.H. Muders, A. Dubrovska, A role for cancer stem cells in therapy resistance: Cellular and molecular mechanisms. Semin. Cancer Biol. 31, 16–27 (2015). https://doi.org/10.1016/j.semcancer.2014.06.004
H.J. Lee, C.F. Li, D. Ruan, J. He, E.D. Montal, S. Lorenz, G.D. Girnun, C.H. Chan, Non-proteolytic ubiquitination of hexokinase 2 by HectH9 controls tumor metabolism and cancer stem cell expansion. Nat. Commun. 10(1), 2625 (2019). https://doi.org/10.1038/s41467-019-10374-y
S. Zhou, J. Peng, L. Xiao, C. Zhou, Y. Fang, Q. Ou, J. Qin, M. Liu, Z. Pan, Z. Hou, TRIM25 regulates oxaliplatin resistance in colorectal cancer by promoting EZH2 stability. Cell Death Dis. 12(5), 463 (2021). https://doi.org/10.1038/s41419-021-03734-4
B.B. Chen, J.R. Glasser, T.A. Coon, C. Zou, H.L. Miller, M. Fenton, J.F. McDyer, M. Boyiadzis, R.K. Mallampalli, F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation. Blood 119(13), 3132–3141 (2012). https://doi.org/10.1182/blood-2011-06-358911
B. Wang, Q. Wang, Z. Wang, J. Jiang, S.C. Yu, Y.F. Ping, J. Yang, S.L. Xu, X.Z. Ye, C. Xu, L. Yang, C. Qian, J.M. Wang, Y.H. Cui, X. Zhang, X.W. Bian, Metastatic consequences of immune escape from NK cell cytotoxicity by human breast cancer stem cells. Cancer Res. 74(20), 5746–5757 (2014). https://doi.org/10.1158/0008-5472.CAN-13-2563
J. Liu, Y. Cheng, M. Zheng, B. Yuan, Z. Wang, X. Li, J. Yin, M. Ye, Y. Song, Targeting the ubiquitination/deubiquitination process to regulate immune checkpoint pathways. Signal 6(1), 28 (2021). https://doi.org/10.1038/s41392-020-00418-x
C. Lyle, S. Richards, K. Yasuda, M.A. Napoleon, J. Walker, N. Arinze, M. Belghasem, I. Vellard, W. Yin, J.D. Ravid, E. Zavaro, R. Amraei, J. Francis, U. Phatak, I.R. Rifkin, N. Rahimi, V.C. Chitalia, C-Cbl targets PD-1 in immune cells for proteasomal degradation and modulates colorectal tumor growth. Sci. Rep. 9(1), 20257 (2019). https://doi.org/10.1038/s41598-019-56208-1
S.C. Casey, L. Tong, Y. Li, R. Do, S. Walz, K.N. Fitzgerald, A.M. Gouw, V. Baylot, I. Gütgemann, M. Eilers, D.W. Felsher, MYC regulates the antitumor immune response through CD47 and PD-L1. Science (New York, N.Y.) 352(6282), 227–231 (2016). https://doi.org/10.1126/science.aac9935
Y. Bei, N. Cheng, T. Chen, Y. Shu, Y. Yang, N. Yang, X. Zhou, B. Liu, J. Wei, Q. Liu, W. Zheng, W. Zhang, H. Su, W.G. Zhu, J. Ji, P. Shen, CDK5 inhibition abrogates TNBC stem-cell property and enhances anti-PD-1 therapy. Adv. Sci. (Weinheim, Baden-Wurttemberg, Germany) 7(22), 2001417 (2020). https://doi.org/10.1002/advs.202001417
P. Zhang, S. Elabd, S. Hammer, V. Solozobova, H. Yan, F. Bartel, S. Inoue, T. Henrich, J. Wittbrodt, F. Loosli, G. Davidson, C. Blattner, TRIM25 has a dual function in the p53/Mdm2 circuit. Oncogene 34(46), 5729–5738 (2015). https://doi.org/10.1038/onc.2015.21
X. Meng, X. Liu, X. Guo, S. Jiang, T. Chen, Z. Hu, H. Liu, Y. Bai, M. Xue, R. Hu, S.C. Sun, X. Liu, P. Zhou, X. Huang, L. Wei, W. Yang, C. Xu, FBXO38 mediates PD-1 ubiquitination and regulates anti-tumour immunity of T cells. Nature 564(7734), 130–135 (2018). https://doi.org/10.1038/s41586-018-0756-0
X.A. Zhou, J. Zhou, L. Zhao, G. Yu, J. Zhan, C. Shi, R. Yuan, Y. Wang, C. Chen, W. Zhang, D. Xu, Y. Ye, W. Wang, Z. Shen, W. Wang, J. Wang, KLHL22 maintains PD-1 homeostasis and prevents excessive T cell suppression. Proc. Natl. Acad. Sci. U. S. A. 117(45), 28239–28250 (2020). https://doi.org/10.1073/pnas.2004570117
J. Zhang, X. Bu, H. Wang, Y. Zhu, Y. Geng, N.T. Nihira, Y. Tan, Y. Ci, F. Wu, X. Dai, J. Guo, Y.H. Huang, C. Fan, S. Ren, Y. Sun, G.J. Freeman, P. Sicinski, W. Wei, Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553(7686), 91–95 (2018). https://doi.org/10.1038/nature25015
Y. Wu, C. Zhang, X. Liu, Z. He, B. Shan, Q. Zeng, Q. Zhao, H. Zhu, H. Liao, X. Cen, X. Xu, M. Zhang, T. Hou, Z. Wang, H. Yan, S. Yang, Y. Sun, Y. Chen, R. Wu, T. Xie, … H. Xia, ARIH1 signaling promotes anti-tumor immunity by targeting PD-L1 for proteasomal degradation. Nat. Commun. 12(1), 2346 (2021). https://doi.org/10.1038/s41467-021-22467-8
X. Huang, V.M. Dixit, Drugging the undruggables: Exploring the ubiquitin system for drug development. Cell Res. 26(4), 484–498 (2016). https://doi.org/10.1038/cr.2016.31
K. Bielskienė, L. Bagdonienė, J. Mozūraitienė, B. Kazbarienė, E. Janulionis, E3 ubiquitin ligases as drug targets and prognostic biomarkers in melanoma. Medicina (Kaunas, Lithuania) 51(1), 1–9 (2015). https://doi.org/10.1016/j.medici.2015.01.007
W. Zhang, Z. Ren, L. Jia, X. Li, X. Jia, Y. Han, Fbxw7 and Skp2 regulate stem cell switch between quiescence and mitotic division in lung adenocarcinoma. Biomed. Res. Int. 2019, 9648269 (2019). https://doi.org/10.1155/2019/9648269
J. Liu, Y. Peng, L. Shi, L. Wan, H. Inuzuka, J. Long, J. Guo, J. Zhang, M. Yuan, S. Zhang, X. Wang, J. Gao, X. Dai, S. Furumoto, L. Jia, P.P. Pandolfi, J.M. Asara, W.G. Kaelin, J. Liu, W. Wei, Skp2 dictates cell cycle-dependent metabolic oscillation between glycolysis and TCA cycle. Cell Res. 31(1), 80–93 (2021). https://doi.org/10.1038/s41422-020-0372-z
W. Jiang, M. Lin, Z. Wang, Dioscin: A new potential inhibitor of Skp2 for cancer therapy. EBioMedicine 51, 102593 (2020). https://doi.org/10.1016/j.ebiom.2019.12.002
E. Malek, M.A. Abdel-Malek, S. Jagannathan, N. Vad, R. Karns, A.G. Jegga, A. Broyl, M. van Duin, P. Sonneveld, F. Cottini, K.C. Anderson, J.J. Driscoll, Pharmacogenomics and chemical library screens reveal a novel SCFSKP2 inhibitor that overcomes Bortezomib resistance in multiple myeloma. Leukemia 31(3), 645–653 (2017). https://doi.org/10.1038/leu.2016.258
J.G. Quirit, S.N. Lavrenov, K. Poindexter, J. Xu, C. Kyauk, K.A. Durkin, I. Aronchik, T. Tomasiak, Y.A. Solomatin, M.N. Preobrazhenskaya, G.L. Firestone, Indole-3-carbinol (I3C) analogues are potent small molecule inhibitors of NEDD4-1 ubiquitin ligase activity that disrupt proliferation of human melanoma cells. Biochem. Pharmacol. 127, 13–27 (2017). https://doi.org/10.1016/j.bcp.2016.12.007
J. Zhang, J.J. Xie, S.J. Zhou, J. Chen, Q. Hu, J.X. Pu, J.L. Lu, Diosgenin inhibits the expression of NEDD4 in prostate cancer cells. Am. J. Transl. Res. 11(6), 3461–3471 (2019)
M. Huang, Y. Zhou, D. Duan, C. Yang, Z. Zhou, F. Li, Y. Kong, Y.C. Hsieh, R. Zhang, W. Ding, W. Xiao, P. Puno, C. Chen, Targeting ubiquitin conjugating enzyme UbcH5b by a triterpenoid PC3-15 from Schisandra plants sensitizes triple-negative breast cancer cells to lapatinib. Cancer Lett. 504, 125–136 (2021). https://doi.org/10.1016/j.canlet.2021.02.009
A. Andrews, K. Warner, C. Rodriguez-Ramirez, A.T. Pearson, F. Nör, Z. Zhang, S. Kerk, A. Kulkarni, J.I. Helman, J.C. Brenner, M.S. Wicha, S. Wang, J.E. Nör, Ablation of Cancer stem cells by therapeutic inhibition of the MDM2-p53 interaction in Mucoepidermoid carcinoma. Clin. Cancer Res. 25(5), 1588–1600 (2019). https://doi.org/10.1158/1078-0432.CCR-17-2730
S. Daniele, B. Costa, E. Zappelli, E. Da Pozzo, S. Sestito, G. Nesi, P. Campiglia, L. Marinelli, E. Novellino, S. Rapposelli, C. Martini, Combined inhibition of AKT/mTOR and MDM2 enhances glioblastoma Multiforme cell apoptosis and differentiation of cancer stem cells. Sci. Rep. 5, 9956 (2015). https://doi.org/10.1038/srep09956
Z. Wei, Y. Liu, Y. Wang, Y. Zhang, Q. Luo, X. Man, F. Wei, X. Yu, Downregulation of Foxo3 and TRIM31 by miR-551b in side population promotes cell proliferation, invasion, and drug resistance of ovarian cancer. Med.l Oncol. (Northwood, London, England) 33(11), 126 (2016). https://doi.org/10.1007/s12032-016-0842-9
Z. Wang, L. Kang, H. Zhang, Y. Huang, L. Fang, M. Li, P.J. Brown, C.H. Arrowsmith, J. Li, J. Wong, AKT drives SOX2 overexpression and cancer cell stemness in esophageal cancer by protecting SOX2 from UBR5-mediated degradation. Oncogene 38(26), 5250–5264 (2019). https://doi.org/10.1038/s41388-019-0790-x
X. Zhang, D. Thummuri, Y. He, X. Liu, P. Zhang, D. Zhou, G. Zheng, Utilizing PROTAC technology to address the on-target platelet toxicity associated with inhibition of BCL-XL. Chem. Commun. (Camb.) 55(98), 14765–14768 (2019). https://doi.org/10.1039/c9cc07217a
X. Li, Y. Song, Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. J. Hematol. Oncol. 13(1), 50 (2020). https://doi.org/10.1186/s13045-020-00885-3
W. Farnaby, M. Koegl, M.J. Roy, C. Whitworth, E. Diers, N. Trainor, D. Zollman, S. Steurer, J. Karolyi-Oezguer, C. Riedmueller, T. Gmaschitz, J. Wachter, C. Dank, M. Galant, B. Sharps, K. Rumpel, E. Traxler, T. Gerstberger, R. Schnitzer, O. Petermann, … A. Ciulli, BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 15(7), 672–680 (2019). https://doi.org/10.1038/s41589-019-0294-6
H. Liao, X. Li, L. Zhao, Y. Wang, X. Wang, Y. Wu, X. Zhou, W. Fu, L. Liu, H.G. Hu, Y.G. Chen, A PROTAC peptide induces durable β-catenin degradation and suppresses Wnt-dependent intestinal cancer. Cell Discov. 6, 35 (2020). https://doi.org/10.1038/s41421-020-0171-1
M.D.M. Noblejas-López, C. Nieto-Jimenez, M. Burgos, M. Gómez-Juárez, J.C. Montero, A. Esparís-Ogando, A. Pandiella, E.M. Galán-Moya, A. Ocaña, Activity of BET-proteolysis targeting chimeric (PROTAC) compounds in triple negative breast cancer. J. Exp. Clin. Cancer Res.: CR 38(1), 383 (2019). https://doi.org/10.1186/s13046-019-1387-5
T. Minko, Nanoformulation of BRD4-degrading PROTAC: Improving Druggability to target the 'Undruggable' MYC in pancreatic Cancer. Trends Pharmacol. Sci. 41(10), 684–686 (2020). https://doi.org/10.1016/j.tips.2020.08.008
J. Hines, S. Lartigue, H. Dong, Y. Qian, C.M. Crews, MDM2-recruiting PROTAC offers superior, synergistic Antiproliferative activity via simultaneous degradation of BRD4 and stabilization of p53. Cancer Res. 79(1), 251–262 (2019). https://doi.org/10.1158/0008-5472.CAN-18-2918
A.C. Qin, H. Jin, Y. Song, Y. Gao, Y.F. Chen, L.N. Zhou, S.S. Wang, X.S. Lu, The therapeutic effect of the BRD4-degrading PROTAC A1874 in human colon cancer cells. Cell Death Dis. 11(9), 805 (2020). https://doi.org/10.1038/s41419-020-03015-6
R. Hu, W.L. Wang, Y.Y. Yang, X.T. Hu, Q.W. Wang, W.Q. Zuo, Y. Xu, Q. Feng, N.Y. Wang, Identification of a selective BRD4 PROTAC with potent antiproliferative effects in AR-positive prostate cancer based on a dual BET/PLK1 inhibitor. Eur. J. Med. Chem. 227, 113922 (2022). https://doi.org/10.1016/j.ejmech.2021.113922
L. Chen, L. Han, S. Mao, P. Xu, X. Xu, R. Zhao, Z. Wu, K. Zhong, G. Yu, X. Wang, Discovery of A031 as effective proteolysis targeting chimera (PROTAC) androgen receptor (AR) degrader for the treatment of prostate cancer. Eur. J. Med. Chem. 216, 113307 (2021). https://doi.org/10.1016/j.ejmech.2021.113307
C. Xu, F. Meng, K.S. Park, A.J. Storey, W. Gong, Y.H. Tsai, E. Gibson, S.D. Byrum, D. Li, R.D. Edmondson, S.G. Mackintosh, M. Vedadi, L. Cai, A.J. Tackett, H.Ü. Kaniskan, J. Jin, G.G. Wang, A NSD3-targeted PROTAC suppresses NSD3 and cMyc oncogenic nodes in cancer cells. Cell Chem. Biol. 29(3), 386–397.e9 (2022). https://doi.org/10.1016/j.chembiol.2021.08.004
Y. Li, J. Yang, A. Aguilar, D. McEachern, S. Przybranowski, L. Liu, C.Y. Yang, M. Wang, X. Han, S. Wang, Discovery of MD-224 as a first-in-class, highly potent, and efficacious proteolysis targeting chimera murine double minute 2 degrader capable of achieving complete and durable tumor regression. J. Med. Chem. 62(2), 448–466 (2019). https://doi.org/10.1021/acs.jmedchem.8b00909
J. Yang, Y. Li, A. Aguilar, Z. Liu, C.Y. Yang, S. Wang, Simple structural modifications converting a Bona fide MDM2 PROTAC degrader into a molecular glue molecule: A cautionary tale in the design of PROTAC degraders. J. Med. Chem. 62(21), 9471–9487 (2019). https://doi.org/10.1021/acs.jmedchem.9b00846
B. Wang, S. Wu, J. Liu, K. Yang, H. Xie, W. Tang, Development of selective small molecule MDM2 degraders based on nutlin. Eur. J. Med. Chem. 176, 476–491 (2019). https://doi.org/10.1016/j.ejmech.2019.05.046
D. Ma, Y. Zou, Y. Chu, Z. Liu, G. Liu, J. Chu, M. Li, J. Wang, S.Y. Sun, Z. Chang, A cell-permeable peptide-based PROTAC against the oncoprotein CREPT proficiently inhibits pancreatic cancer. Theranostics 10(8), 3708–3721 (2020). https://doi.org/10.7150/thno.41677
K. Wang, H. Zhou, Proteolysis targeting chimera (PROTAC) for epidermal growth factor receptor enhances anti-tumor immunity in non-small cell lung cancer. Drug Dev. Res. 82(3), 422–429 (2021). https://doi.org/10.1002/ddr.21765
Q. Liu, G. Tu, Y. Hu, Q. Jiang, J. Liu, S. Lin, Z. Yu, G. Li, X. Wu, Y. Tang, X. Huang, J. Xu, Y. Liu, L. Wu, Discovery of BP3 as an efficacious proteolysis targeting chimera (PROTAC) degrader of HSP90 for treating breast cancer. Eur. J. Med. Chem. 228, 114013 (2022). https://doi.org/10.1016/j.ejmech.2021.114013
L. Zhang, Q. Liu, K.W. Liu, Z.Y. Qin, G.X. Zhu, L.T. Shen, N. Zhang, B.Y. Liu, L.R. Che, J.Y. Li, T. Wang, L.Z. Wen, K.J. Liu, Y. Guo, X.R. Yin, X.W. Wang, Z.H. Zhou, H.L. Xiao, Y.H. Cui, X.W. Bian, … B. Wang, SHARPIN stabilizes β-catenin through a linear ubiquitination-independent manner to support gastric tumorigenesis. Gastric Cancer 24(2), 402–416 (2021). https://doi.org/10.1007/s10120-020-01138-5
Z.Y. Qin, T. Wang, S. Su, L.T. Shen, G.X. Zhu, Q. Liu, L. Zhang, K.W. Liu, Y. Zhang, Z.H. Zhou, X.N. Zhang, L.Z. Wen, Y.L. Yao, W.J. Sun, Y. Guo, K.J. Liu, L. Liu, X.W. Wang, Y.L. Wei, J. Wang, … B. Wang, BRD4 promotes gastric Cancer progression and metastasis through acetylation-dependent stabilization of Snail. Cancer Res. 79(19), 4869–4881 (2019). https://doi.org/10.1158/0008-5472.CAN-19-0442
C.Y. Fong, O. Gilan, E.Y. Lam, A.F. Rubin, S. Ftouni, D. Tyler, K. Stanley, D. Sinha, P. Yeh, J. Morison, G. Giotopoulos, D. Lugo, P. Jeffrey, S.C. Lee, C. Carpenter, R. Gregory, R.G. Ramsay, S.W. Lane, O. Abdel-Wahab, T. Kouzarides, … M.A. Dawson, BET inhibitor resistance emerges from leukaemia stem cells. Nature 525(7570), 538–542 (2015). https://doi.org/10.1038/nature14888
J. Ding, N. Sharon, Z. Bar-Joseph, Temporal modelling using single-cell transcriptomics. Nat. Rev. Genet. 23(6), 355–368 (2022). https://doi.org/10.1038/s41576-021-00444-7
X. Wei, S. Fu, H. Li, Y. Liu, S. Wang, W. Feng, Y. Yang, X. Liu, Y.Y. Zeng, M. Cheng, Y. Lai, X. Qiu, L. Wu, N. Zhang, Y. Jiang, J. Xu, X. Su, C. Peng, L. Han, W.P. Lou, … Y. Gu, Single-cell Stereo-seq reveals induced progenitor cells involved in axolotl brain regeneration. Science (New York, N.Y.). 377(6610), eabp9444 (2022). https://doi.org/10.1126/science.abp9444
Acknowledgements
We thank all the members in the laboratory for helpful discussions on the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (No. 2022YFA1105300 to Bin Wang), National Natural Science Foundation of China (NSFC Nos. 81872027, 91959111, and 81822032 to Bin Wang), Natural Science Foundation of Chongqing (No. CSTC2019JCYJJQX0027 to Bin Wang), Medical Scientific Research Project of Chongqing Medical and Health Committee (2018GDRC006 to Yi Zhang), Funding from the Jin Feng laboratory to Bin Wang, and Funding from the Army Medical University (Nos. 2019CXLCA001, 2018XLC2023, and 2019XQY19 to Bin Wang), the National Institutes of Health, USA (R01AG077574), Nebraska Department of Health & Human Services (DHHS) LB595, LB606, and Creighton University startup funds (LB692) to Brian North. Through LB595, LB606 and LB692, this work is supported by revenue from Nebraska’s excise tax on cigarettes awarded to Brian North of Creighton University through the Nebraska Department of Health & Human Services (DHHS). Its contents represent the view(s) of the author(s) and do not necessarily represent the official views of the State of Nebraska or DHHS.
Author information
Authors and Affiliations
Contributions
Qiang Zou with the assistance of Meng Liu collected all the reference, made the table, and wrote the manuscript draft. Qiang Zou and Yi Zhang designed and drawn all the figures. Brian J. North checked the structure and grammar. Bin Wang discussed and amended the manuscript. Bin Wang, Brian J. North and Yi Zhang supervised the whole process. Bin Wang and Brian J. North edited the manuscript and approved the submission. All the authors contributed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethical approval
This study was approved by the Medical Ethics Committee of Army Medical University.
Competing interests
The authors have declared no potential conflicts of interest in this review.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qiang Zou and Meng Liu share co-first authorship.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zou, Q., Liu, M., Liu, K. et al. E3 ubiquitin ligases in cancer stem cells: key regulators of cancer hallmarks and novel therapeutic opportunities. Cell Oncol. 46, 545–570 (2023). https://doi.org/10.1007/s13402-023-00777-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-023-00777-x