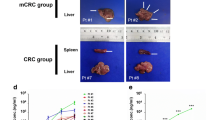
Abstract
Purpose
We aimed to elucidate the applicability of tumor organoids for inherent drug resistance of primary liver cancer (PLC) and mechanisms of acquired drug resistance.
Methods
PLC tissues were used to establish organoids, organoid-derived xenograft (ODX) and patient-derived xenograft (PDX) models. Acquired drug resistance was induced in hepatocellular carcinoma (HCC) organoids. Gene expression profiling was performed by RNA-sequencing.
Results
Fifty-two organoids were established from 153 PLC patients. Compared with establishing PDX models, establishing organoids of HCC showed a trend toward a higher success rate (29.0% vs. 23.7%) and took less time (13.0 ± 4.7 vs. 25.1 ± 5.4 days, p = 2.28 × 10−13). Larger tumors, vascular invasion, higher serum AFP levels, advanced stages and upregulation of stemness- and proliferation-related genes were significantly associated with the successful establishment of HCC organoids and PDX. Organoids and ODX recapitulated PLC histopathological features, but were enriched in more aggressive cell types. PLC organoids were mostly resistant to lenvatinib in vitro but sensitive to lenvatinib in ODX models. Stemness– and epithelial–mesenchymal transition (EMT)–related gene sets were found to be upregulated, whereas liver development– and liver specific molecule–related gene sets were downregulated in acquired sorafenib-resistant organoids. Targeting the mTOR signaling pathway was effective in treating acquired sorafenib-resistant HCC organoids, possibly via inducing phosphorylated S6 kinase. Genes upregulated in acquired sorafenib-resistant HCC organoids were associated with an unfavorable prognosis.
Conclusions
HCC organoids perform better than PDX for drug screening. Acquired sorafenib resistance in organoids promotes HCC aggressiveness via facilitating stemness, retro-differentiation and EMT. Phosphorylated S6 kinase may be predictive for drug resistance in HCC.







Similar content being viewed by others
Data availability
The original RNA-Seq data of tumor tissues are deposited in the Sequence Read Archive (SRA) database with accession PRJNA841062. The original RNA-Seq data in this study are deposited in the Gene Expression Omnibus (GEO) database with accession GSE182593. Other data are available from the corresponding author upon reasonable request.
References
H. Sung, J. Ferlay, R.L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal, F. Bray, Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021)
D. Jiang, L. Zhang, W. Liu, Y. Ding, J. Yin, R. Ren, Q. Li, Y. Chen, J. Shen, X. Tan, H. Zhang, G. Cao, Trends in cancer mortality in China from 2004 to 2018: A nationwide longitudinal study. Cancer Commun. (Lond.) 41, 1024–1036 (2021)
F. Yang, L. Ma, Y. Yang, W. Liu, J. Zhao, X. Chen, M. Wang, H. Zhang, S. Cheng, F. Shen, H. Wang, W. Zhou, G. Cao, Contribution of hepatitis B virus infection to the aggressiveness of primary liver cancer: A clinical epidemiological study in eastern China. Front. Oncol. 9, 370 (2019)
G. Marasco, F. Ravaioli, A. Vestito, B. Rossini, E. Dajti, L. Colecchia, K. Gjini, M. Renzulli, R. Golfieri, D. Festi, A. Colecchia, Predictive factors for hepatocellular carcinoma recurrence after curative treatments. Hepatoma Res. 6, 33 (2020)
A. Rammohan, M. Rela, Risk factors and management of post-liver transplant recurrence of hepatocellular carcinoma. Hepatoma Res. 7, 49 (2021)
Y. Lai, X. Wei, S. Lin, L. Qin, L. Cheng, P. Li, Current status and perspectives of patient-derived xenograft models in cancer research. J. Hematol. Oncol. 10, 106 (2017)
M.R. Kuracha, P. Thomas, B.W. Loggie, V. Govindarajan, Patient-derived xenograft mouse models of pseudomyxoma peritonei recapitulate the human inflammatory tumor microenvironment. Cancer Med. 5, 711–719 (2016)
G.J. Yoshida, Applications of patient-derived tumor xenograft models and tumor organoids. J. Hematol. Oncol. 7, 13:4 (2020)
S. Shi, M.M.A. Verstegen, H.P. Roest, A.I. Ardisasmita, W. Cao, F.J.M. Roos, P.E. de Ruiter, M. Niemeijer, Q. Pan, J.N.M. IJzermans, L.J.W. van der Laan, Recapitulating cholangiopathy-associated necroptotic cell death in vitro using human cholangiocyte organoids. Cell Mol. Gastroenterol. Hepatol. 13, 541–564 (2021)
L. Broutier, G. Mastrogiovanni, M.M. Verstegen, H.E. Francies, L.M. Gavarró, C.R. Bradshaw, G.E. Allen, R. Arnes-Benito, O. Sidorova, M.P. Gaspersz, N. Georgakopoulos, B.K. Koo, S. Dietmann, S.E. Davies, R.K. Praseedom, R. Lieshout, J.N.M. Ijzermans, S.J. Wigmore, K. Saeb-Parsy, et al., Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 23, 1424–1435 (2017)
S. Nuciforo, I. Fofana, M.S. Matter, T. Blumer, D. Calabrese, T. Boldanova, S. Piscuoglio, S. Wieland, F. Ringnalda, G. Schwank, L.M. Terracciano, C.K.Y. Ng, M.H. Heim, Organoid models of human liver cancers derived from tumor needle biopsies. Cell Rep. 24, 1363–1376 (2018)
L. Li, H. Knutsdottir, K. Hui, M.J. Weiss, J. He, B. Philosophe, A.M. Cameron, C.L. Wolfgang, T.M. Pawlik, G. Ghiaur, A.J. Ewald, E. Mezey, J.S. Bader, F.M. Selaru, Human primary liver cancer organoids reveal intratumor and interpatient drug response heterogeneity. JCI Insight 4, e121490 (2019)
S. Wang, Y. Wang, X. Xun, C. Zhang, X. Xiang, Q. Cheng, S. Hu, Z. Li, J. Zhu, Hedgehog signaling promotes sorafenib resistance in hepatocellular carcinoma patient-derived organoids. J. Exp. Clin. Cancer Res. 39, 22 (2020)
Y. Zhao, Z.X. Li, Y.J. Zhu, J. Fu, X.F. Zhao, Y.N. Zhang, S. Wang, J.M. Wu, K.T. Wang, S.C.J. Wu, S.Y. Shen, X. Wu, H.Y. Wang, D. Gao, L. Chen, Single cell transcriptome analysis uncovers intratumoral heterogeneity and underlying mechanisms for drug resistance in hepatobiliary tumor organoids. Adv. Sci, (Weinh.) 8, e2003897 (2021)
C.O.N. Leung, M. Tong, K.P.S. Chung, L. Zhou, N. Che, K.H. Tang, J. Ding, E.Y.T. Lau, I.O.L. Ng, S. Ma, T.K.W. Lee, Overriding adaptive resistance to sorafenib through combination therapy with Src homology 2 domain-containing phosphatase 2 blockade in hepatocellular carcinoma. Hepatology 72, 155–168 (2020)
S.R. Veiga, X. Ge, C.A. Mercer, M.I. Hernández-Álvarez, H.E. Thomas, J. Hernandez-Losa, R.Y. Cajal, S, Zorzano A, Thomas G, Kozma SC., Phenformin-induced mitochondrial dysfunction sensitizes hepatocellular carcinoma for dual inhibition of mTOR. Clin. Cancer Res. 24, 3767–3780 (2018)
W. Chang, X. Gao, Y. Han, Y. Du, Q. Liu, L. Wang, X. Tan, Q. Zhang, Y. Liu, Y. Zhu, Y. Yu, X. Fan, H. Zhang, W. Zhou, J. Wang, C. Fu, G. Cao, Gene expression profiling-derived immunohistochemistry signature with high prognostic value in colorectal carcinoma. Gut 63, 1457–1467 (2014)
M.A. Rodríguez-Hernández, R. Chapresto-Garzón, M. Cadenas, E. Navarro-Villarán, M. Negrete, M.A. Gómez-Bravo, V.M. Victor, F.J. Padillo, J. Muntané, Differential effectiveness of tyrosine kinase inhibitors in 2D/3D culture according to cell differentiation, p53 status and mitochondrial respiration in liver cancer cells. Cell Death Dis. 11, 339 (2020)
A.M. Bolger, M. Lohse, B. Usadel, Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–21120 (2014)
R.K. Patel, M. Jain, NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PLoS One 7, e30619 (2012)
D. Kim, B. Langmead, S.L. Salzberg, HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015)
M. Pertea, G.M. Pertea, C.M. Antonescu, T.C. Chang, J.T. Mendell, S.L. Salzberg, StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015)
C. Trapnell, A. Roberts, L. Goff, G. Pertea, D. Kim, D.R. Kelley, H. Pimentel, S.L. Salzberg, J.L. Rinn, L. Pachter, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 7, 562–578 (2012)
B. Langmead, S.L. Salzberg, Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359 (2012)
A. Roberts, L. Pachter, Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods 10, 71–73 (2013)
D.J. McCarthy, Y. Chen, G.K. Smyth, Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012)
A. Subramanian, P. Tamayo, V.K. Mootha, S. Mukherjee, B.L. Ebert, M.A. Gillette, A. Paulovich, S.L. Pomeroy, T.R. Golub, E.S. Lander, J.P. Mesirov, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550 (2005)
F.M. Ippen, C.A. Alvarez-Breckenridge, B.M. Kuter, A.L. Fink, I.V. Bihun, M. Lastrapes, T. Penson, S.P. Schmidt, G.R. Wojtkiewicz, J. Ning, M. Subramanian, A. Giobbie-Hurder, M. Martinez-Lage, S.L. Carter, D.P. Cahill, H. Wakimoto, P.K. Brastianos, The dual PI3K/mTOR pathway inhibitor GDC-0084 achieves antitumor activity in PIK3CA-mutant breast cancer brain metastases. Clin. Cancer Res. 25, 3374–3383 (2019)
N.Y. Kim, S. Lee, J. Yu, N. Kim, S.S. Won, H. Park, W.D. Heo, Optogenetic control of mRNA localization and translation in live cells. Nat. Cell Biol. 22, 341–352 (2020)
J. Yin, N. Li, Y. Han, J. Xue, Y. Deng, J. Shi, W. Guo, H. Zhang, H. Wang, S. Cheng, G. Cao, Effect of antiviral treatment with nucleotide/nucleoside analogs on postoperative prognosis of hepatitis B virus-related hepatocellular carcinoma: A two-stage longitudinal clinical study. J. Clin. Oncol. 31, 3647–3655 (2013)
T. Kawai, K. Yasuchika, T. Ishii, H. Katayama, E.Y. Yoshitoshi, S. Ogiso, S. Kita, K. Yasuda, K. Fukumitsu, M. Mizumoto, E. Hatano, S. Uemoto, Keratin 19, a cancer stem cell marker in human epatocellular carcinoma. Clin. Cancer Res. 21, 3081–3091 (2015)
G. Chen, Y. Wang, X. Zhao, X.Z. Xie, J.G. Zhao, T. Deng, Z.Y. Chen, H.B. Chen, Y.F. Tong, Z. Yang, X.W. Ding, P.Y. Guo, H.T. Yu, L.J. Wu, S.N. Zhang, Q.D. Zhu, J.J. Li, Y.F. Shan, F.X. Yu, et al., A positive feedback loop between Periostin and TGFβ1 induces and maintains the stemness of hepatocellular carcinoma cells via AP-2α activation. J. Exp. Clin. Cancer Res. 40, 218 (2021)
Y. Pan, M. Han, X. Zhang, Y. He, C. Yuan, Y. Xiong, X. Li, C. Zeng, K. Lu, H. Zhu, X. Lu, Q. Liu, H. Liang, Z. Liao, Z. Ding, Z. Zhang, X. Chen, W. Zhang, B. Zhang, Discoidin domain receptor 1 promotes hepatocellular carcinoma progression through modulation of SLC1A5 and the mTORC1 signaling pathway. Cell. Oncol. 45, 163–178 (2022)
D.J. Marchant, C.L. Bellac, T.J. Moraes, S.J. Wadsworth, A. Dufour, G.S. Butler, L.M. Bilawchuk, R.G. Hendry, A.G. Robertson, C.T. Cheung, J. Ng, L. Ang, Z. Luo, K. Heilbron, M.J. Norris, W. Duan, T. Bucyk, A. Karpov, L. Devel, et al., A new transcriptional role for matrix metalloproteinase-12 in antiviral immunity. Nat. Med. 20, 493–502 (2014)
Q. Wu, W. Zhou, S. Yin, Y. Zhou, T. Chen, J. Qian, R. Su, L. Hong, H. Lu, F. Zhang, H. Xie, L. Zhou, S. Zheng, Blocking triggering receptor expressed on myeloid cells-1-positive tumor-associated macrophages induced by hypoxia reverses immunosuppression and anti-programmed cell death ligand 1 resistance in liver cancer. Hepatology 70, 198–214 (2019)
J. Shen, T. Liu, J. Lv, S. Xu, Identification of an immune-related prognostic gene CLEC5A based on immune microenvironment and risk modeling of ovarian cancer. Front Cell Dev. Biol. 9, 746932 (2021)
Y.H. Kuo, Y.H. Yen, Y.Y. Chen, K.M. Kee, C.H. Hung, S.N. Lu, T.H. Hu, C.H. Chen, J.H. Wang, Nivolumab versus regorafenib in patients with hepatocellular carcinoma after sorafenib failure. Front. Oncol. 11, 683341 (2021)
M. Zhu, L. Li, T. Lu, H. Yoo, J. Zhu, P. Gopal, S.C. Wang, M.R. Porembka, N.E. Rich, S. Kagan, M. Odewole, V. Renteria, A.K. Waljee, T. Wang, A.G. Singal, A.C. Yopp, H. Zhu, Uncovering biological factors that regulate hepatocellular carcinoma growth using patient-derived xenograft assays. Hepatology 72, 1085–1101 (2020)
J.L. Carroll, L.L. Nielsen, S.B. Pruett, J.M. Mathis, The role of natural killer cells in adenovirus-mediated p53 gene therapy. Mol. Cancer Ther. 1, 49–60 (2001)
Q. Zhang, H. Liu, H. Wang, M. Lu, Y. Miao, J. Ding, H. Li, X. Gao, S. Sun, J. Zheng, Lenvatinib promotes antitumor immunity by enhancing the tumor infiltration and activation of NK cells. Am. J. Cancer Res. 9, 1382–1395 (2019)
J.M. Llovet, S. Ricci, V. Mazzaferro, P. Hilgard, E. Gane, J.F. Blanc, A.C. de Oliveira, A. Santoro, J.L. Raoul, A. Forner, M. Schwartz, C. Porta, S. Zeuzem, L. Bolondi, T.F. Greten, P.R. Galle, J.F. Seitz, I. Borbath, D. Häussinger, et al., SHARP investigators study group. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390 (2008)
Q. Zhu, H. Ren, X. Li, B. Qian, S. Fan, F. Hu, L. Xu, B. Zhai, Silencing KIF14 reverses acquired resistance to sorafenib in hepatocellular carcinoma. Aging (Albany NY) 12, 22975–23003 (2020)
V. Tovar, H. Cornella, A. Moeini, S. Vidal, Y. Hoshida, D. Sia, J. Peix, L. Cabellos, C. Alsinet, S. Torrecilla, I. Martinez-Quetglas, J.J. Lozano, C. Desbois-Mouthon, M. Solé, J. Domingo-Domenech, A. Villanueva, J.M. Llovet, Tumour initiating cells and IGF/FGF signalling contribute to sorafenib resistance in hepatocellular carcinoma. Gut 66, 530–540 (2017)
M. Liu, Q. Hu, M. Tu, X. Wang, Z. Yang, G. Yang, R. Luo, MCM6 promotes metastasis of hepatocellular carcinoma via MEK/ERK pathway and serves as a novel serum biomarker for early recurrence. J. Exp. Clin. Cancer Res. 37, 10 (2018)
J. Wang, Z. Li, C. Zuo, Q. Xie, H. Li, J. Jia, Z. Zhen, R. Qi, Z. Li, D. Liu, B. Sun, Knockdown of RRS1 by lentiviral-mediated RNAi promotes apoptosis and suppresses proliferation of human hepatocellular carcinoma cells. Oncol. Rep. 38, 2166–2172 (2017)
Z. Zhou, X. Liu, Y. Li, J. Li, W. Deng, J. Zhong, L. Chen, Y. Li, X. Zeng, G. Wang, J. Zhu, B. Fu, TP53INP2 modulates epithelial-to-mesenchymal transition via the GSK-3β/β-catenin/Snail1 pathway in bladder cancer cells. Onco Targets Ther 13, 9587–9597 (2020)
A. Surcel, E.S. Schiffhauer, D.G. Thomas, Q. Zhu, K.T. DiNapoli, M. Herbig, O. Otto, H. West-Foyle, A. Jacobi, M. Kräter, K. Plak, J. Guck, E.M. Jaffee, P.A. Iglesias, R.A. Anders, D.N. Robinson, Targeting mechanoresponsive proteins in pancreatic cancer: 4-hydroxyacetophenone blocks dissemination and invasion by activating MYH14. Cancer Res. 79, 4665–4678 (2019)
B. Zhang, H.Y. Wang, D.X. Zhao, D.X. Wang, Q. Zeng, J.F. Xi, X. Nan, L.J. He, J.N. Zhou, X.T. Pei, W. Yue, The splicing regulatory factor hnRNPU is a novel transcriptional target of c-Myc in hepatocellular carcinoma. FEBS Lett. 595, 68–84 (2021)
S. Talukdar, L. Emdad, S.K. Das, P.B. Fisher, EGFR: An essential receptor tyrosine kinase-regulator of cancer stem cells. Adv. Cancer Res. 147, 161–188 (2020)
J. Chen, H. Zhu, Q. Liu, D. Ning, Z. Zhang, L. Zhang, J. Mo, P. Du, X. Liu, S. Song, Y. Fan, H. Liang, J. Liu, B. Zhang, X. Chen, DEPTOR induces a partial epithelial-to-mesenchymal transition and metastasis via autocrine TGFβ1 signaling and is associated with poor prognosis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 38, 273 (2019)
J. Xing, V. Bhuria, K.C. Bui, M.L.T. Nguyen, Z. Hu, C.J. Hsieh, K. Wittstein, M. Stadler, L. Wilkens, J. Li, M. Kalesse, P. Bozko, R.R. Plentz, Haprolid inhibits tumor growth of hepatocellular carcinoma through Rb/E2F and Akt/mTOR inhibition. Cancers (Basel) 12, 615 (2020)
L. Zhu, N. Yang, G. Du, C. Li, G. Liu, S. Liu, Y. Xu, Y. Di, W. Pan, X. Li, LncRNA CRNDE promotes the epithelial-mesenchymal transition of hepatocellular carcinoma cells via enhancing the Wnt/β-catenin signaling pathway. J. Cell. Biochem. 120, 1156–1164 (2018)
J. Stebbing, A. Filipović, G. Giamas, Claudin-1 as a promoter of EMT in hepatocellular carcinoma. Oncogene 32, 4871–4872 (2013)
M. Hu, M. Li, H. Huang, C. Lu, Isolated cancer stem cells from human liver cancer: Morphological and functional characteristics in primary culture. Clin. Transl. Oncol. 24, 48–56 (2022)
X. Sang, F. Wu, D. Wu, S. Lin, J. Li, N. Zhao, X. Chen, A. Xu, Human hepatic cancer stem cells (HCSCs) markers correlated with immune infiltrates reveal prognostic significance of hepatocellular carcinoma. Front. Genet. 11, 112 (2020)
I. Pastushenko, C. Blanpain, EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29, 212–226 (2019)
I. Pastushenko, A. Brisebarre, A. Sifrim, M. Fioramonti, T. Revenco, S. Boumahdi, A. Van Keymeulen, D. Brown, V. Moers, S. Lemaire, S. De Clercq, E. Minguijón, C. Balsat, Y. Sokolow, C. Dubois, F. De Cock, S. Scozzaro, F. Sopena, A. Lanas, et al., Identification of the tumour transition states occurring during EMT. Nature 556, 463–468 (2018)
M. Masuda, W.Y. Chen, A. Miyanaga, Y. Nakamura, K. Kawasaki, T. Sakuma, M. Ono, C.L. Chen, K. Honda, T. Yamada, Alternative mammalian target of rapamycin (mTOR) signal activation in sorafenib-resistant hepatocellular carcinoma cells revealed by array-based pathway profiling. Mol. Cell. Proteomics 13, 1429–1438 (2014)
I. Eliseeva, M. Vasilieva, L.P. Ovchinnikov, Translation of human β-actin mRNA is regulated by mTOR pathway. Genes (Basel) 10, 96 (2019)
Funding
This work was supported by the National Key Basic Research Program of China [grant number 2015CB554006 (GC)], the National Natural Science Foundation of China [grant numbers 91529305 (GC), 81520108021 (GC), 81673250 (GC), 81521091 (GC), 82003538 (WL), and 81502882 (XC)], the Shanghai Yangfan Program [grant numbers 20YF1458800 (WL)] and the “3-year public health promotion” program of Shanghai Municipal Health Commission [grant numbers GWV-10.1-XK17 (GC)].
Author information
Authors and Affiliations
Contributions
Cao G conceived and supervised the study. Xian L, Zhao P, and Wei Z were responsible for the culture and maintenance of tumor organoids. Chen X performed the bioinformatics analyses. Ji H and Zhao J did surgical treatments and provided suitable clinical specimens. Liu D, Li Z, Liu W, Zhou X, Fan J, Zhu X, Yin J, and Tan X took part in the cell experiments and animal care. Qian Y and Dong H took part in the histology and immunohistochemistry assays. Chen X, Han X, and Yu H conducted the statistical analyses. Cao G interpreted the data and drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
This study protocol was reviewed and approved by Ethics Committee of Eastern Hepatobiliary Surgery Hospital, approval number 2019UE-023. All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki Declaration and its later amendments. Informed consent was obtained from all participants included in the study. All animal experiments in this study were conducted in accordance with the guidelines of the animal ethical committee for animal experimentation in China. The experimental design was approved by the Institutional Animal Care and Use Committee of Second Military Medical University.
Consent for publication
All patients signed informed consent regarding publishing their data.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information

Fig. S1
The success rate and average days of constructing PLC organoids and PDX model. (a) The success rate of establishing tumor organoids of each HCC and ICC. (b) The success rate of establishing tumor PDX models of HCC and ICC. (c) The duration of tumor organoids’ culture from tissue separation to the first passage was 13.0 ± 4.7 and 13.8 ± 3.4 days for HCC and ICC, respectively. (d) The duration of PDX construction from the first tumor implantation to the second implantation was 25.1 ± 5.4 and 33. 0 ± 5.7 days for HCC and ICC, respectively. (PNG 82 kb)

Fig. S2
H&E staining images of organoids at different passages. Histological features of the first generation organoids (fist line) and the seventh generation organoids (second line) of HCC-118, ICC-6, and CHC-3. Scale bars, 50 μm (PNG 1743 kb)

Fig. S3
Quality control of RNA-Seq data derived from HCC samples. The number of read pairs (marked on left Y-axis), rate of high quality (HQ) reads (calculated by NGSQCToolkit) (marked on right Y-axis), and the alignment rate (calculated by HISAT2) (marked on right Y-axis) for each sample were plotted. (PNG 41 kb)

Fig. S4
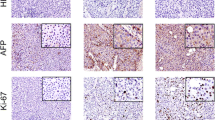
Immunohistochemistry of tumor tissues, organoids, and ODX and PDX from PLC patients of major histotypes. (a) Expression of AFP in tumor tissues, tumor organoids, ODX, and PDX derived from HCC-25, HCC-118, ICC-6, and CHC-3 patients. (b) Expression of CK-19 in tissues, organoids, ODX, and PDX. (c) Expression of EpCAM in tissues, organoids, ODX and PDX. Scale bars, 50 μm. CK19, cytokeratin 19; PLC, primary liver cancer; HCC, hepatocellular carcinoma; ICC, intrahepatic cholangiocarcinoma; CHC, hepatocellular cholangiocarcinoma; PDX, patient-derived xenograft; and ODX, organoids-derived xenograft. (PNG 7243 kb)

Fig. S5
The stemness- and epithelial–mesenchymal transition-related gene sets enriched in each of the four acquired sorafenib-resistant HCC organoids. (a) gene sets enriched in the organoid of HCC-52; (b) gene sets enriched in the organoid of HCC-118; (c) gene sets enriched in the organoid of HCC-10; and (d) gene sets enriched in the organoid of HCC-25. NES, normalized enrichment score; FDR, false discovery rate. (PNG 141 kb)

Fig. S6
The heterogeneity of stemness- and epithelial–mesenchymal transition-related gene expression patterns in four HCC organoids with and without acquired sorafenib resistance. (a) Western blotting showed the expression patterns of N-cadherin, Vimentin, Claudin-1, CD44, ABCG2, and EpCAM in HCC-118, HCC-25, HCC-10, and HCC-52 parental and sorafenib-resistant organoids, respectively. (b) Immunohistochemistry showed the expression of CD44, EpCAM, N-cadherin, and Vimentin in the parental and acquired sorafenib-resistant organoids of HCC-118 and HCC-25. Scale bars, 50 μm. (PNG 1736 kb)

Fig. S7
Association of the differentially expressed genes in acquired sorafenib-resistant HCC organoids with the prognosis (a) overall survival; and (b) recurrence-free survival. (PNG 37 kb)
Table S1
(XLSX 19 kb)
Table S2
(DOCX 21 kb)
Table S3
(DOCX 25 kb)
Table S4
(DOCX 19 kb)
Table S5
(DOCX 22 kb)
Table S6
(DOCX 24 kb)
Table S7
(DOCX 16 kb)
Table S8
(DOCX 59 kb)
Table S9
(DOCX 56 kb)
Table S10
(DOCX 82 kb)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xian, L., Zhao, P., Chen, X. et al. Heterogeneity, inherent and acquired drug resistance in patient-derived organoid models of primary liver cancer. Cell Oncol. 45, 1019–1036 (2022). https://doi.org/10.1007/s13402-022-00707-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-022-00707-3