Abstract
Background
Cervical intraepithelial neoplasia (CIN), a frequently encountered disease caused by Human Papilloma Virus (HPV) is often diagnosed in formaldehyde-fixed paraffin embedded (FFPE) punch biopsies. Since it is known that this procedure strongly affects the water-soluble proteins contained in the cervical tissue we decided to investigate whether a water-soluble protein-saving biopsy processing method can be used to support the diagnosis of normal and CIN.
Methods
Cervical punch biopsies from 55 women were incubated for 24 h at 4°C in RPMI1640 medium for protein analysis prior to usual FFPE processing and p16 and Ki67-supported histologic consensus diagnosis was assessed. The biopsy supernatants were subjected to surface-enhanced laser desorption-ionization time of flight mass spectrometry (SELDI-TOF MS) for identifying differentially expressed proteins. Binary logistic regression and classification and regression trees (CART) were used to develop a classification model.
Results
The age of the patients ranged from 26 to 40 years (median 29.7). The consensus diagnoses were normal cervical tissue (n = 10) and CIN2-3 (n = 45). The mean protein concentration was 1.00 and 1.09 mg/ml in the normal and CIN2-3 group, respectively. The peak detection and clustering process resulted in 40 protein peaks. Many of these peaks differed between the two groups, but only three had independent discriminating power. The overall classification results were 88%.
Conclusions
Water-soluble proteins sampled from punch biopsies are promising to assist the diagnosis of normal and CIN2-3.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Background
Among the cancers affecting women, cervical cancer has the second highest occurrence worldwide. In developing countries where 80% of the deaths caused by this disease occur, it is the cancer type that affects most women [1]. The most important risk factor associated with the development of cervical cancer is the infection of cervical epithelial cells by high-risk Human Papilloma Virus genotype (hr-HPV) [2]. The tumor development can take 10–25 years, during this time the cervical epithelium develops a non-invasive neoplastic lesion called cervical intraepithelial neoplasia (CIN) [3]. The World Health Organization distinguishes three CIN grades (CIN1, CIN2 and CIN3) according to the degree of epithelial abnormality. This can easily give the faulty impression of a solidified sculpture, as if the three CIN grades are static events, whereas in reality a CIN lesion is a dynamic process that can persist and progress but also regress [4]. When CIN2-3 is diagnosed in a histological punch biopsy, definitive therapy by cone excision is usually promptly performed [5]. The reason for this aggressive invasive therapy is that 5–30% of CIN2-3 patients will eventually develop invasive cancer when left untreated. However, at the same time many (10–40%) CIN2-3 s will regress spontaneously [6]. Recently, it has been shown that functional biomarkers such as Ki67, pRb, p53 and cytokeratine 13/14 are helpful in the prediction of regression or not, and the aggregate information provided by these biomarkers exceeds the value of the classic grading system. Consequently, many more CIN-lesions that can either regress or progress could be more accurately treated [7].
Although these methods are very promising, the fixation process of fresh tissues, required for conventional microscopical and many immunohistochemical stains leads to loss of water-soluble proteins that could be of interest. In addition it affects the remaining proteins by introducing different crosslinks that can make it difficult to obtain them for identification after the fixation process [8–11]. Some of the protein modifications introduced by the fixation process may also lead to misidentifications [9]. Cytokines and chemokines are examples of such proteins whose characterization in CIN-lesions could give valuable information about the patient’s immune reaction against the HPV-infection [12–14].
As the HPV-infection induces a local immune response in the cervix that is barely detectable at plasma/serum level [15] it is important to establish a standardized method for collection of a sample that truly represents the cervix microenvironment. In order to achieve this, several studies have been performed using different sample types like cervicovaginal washings [14, 16], cervical mucus [17] and cells supernatant from cytobrush collection [18], employing different analysis technologies. One of these technologies is surface-enhanced laser desorption-ionization time of flight mass spectrometry (SELDI-TOF MS) which is a powerful tool for identifying differentially expressed proteins. These proteins can be potentially diagnostic and prognostic biomarkers in neoplasia [19, 20].
The aim of this study was to develop a protein-saving biopsy processing method with similarities to the method published by Celis et al. [21], and to utilize SELDI-TOF MS to analyze the proteins from the biopsies, while at the same time preserving the tissue for conventional microscopic and immunohistochemical studies. In this pilot study, we will investigate if it is possible to separate normal cervical tissue from CIN2-3 lesions.
2 Methods
2.1 Study population
The current study is a sub-project from a larger prospective study, approved by the Regional Medical Ethics Committee of Helse Vest, Norway, the Norwegian Data Inspection, and the Health Directorate of Norway, #33.06, #17185 and #07/330. Two hundred and fifty-six patients have been enrolled in this prospective study. The criterion for selection in this large prospective study is a cytological abnormal smear followed by cervical biopsy in women with previously normal smears. Of these 256 patients with cervical punch biopsy samples, 45 diagnosed as CIN2-3 (n = 45) were selected. From women in the same age range, biopsies that were histologically and immunohistochemically normal (n = 10) were also used.
2.2 Sample collection
For soluble protein recovery, biopsies of normal and cervical neoplastic epithelium were collected and immediately placed in polystyrene tubes (Sarsted, Nurmbrecht, Germany) containing 5 ml of RPMI1640 culture medium (Gibco, Carslbad, USA) and stored for 24 h at 4°C. Afterwards, the supernatants were collected, split into aliquots of 500 μl and stored at −80°C until analysis.
2.3 Pathology
After 24 h incubation in the RPMI1640 medium, the biopsies were routinely fixed in buffered 4% formaldehyde, embedded in paraffin, cut at 4 μm, and stained with hematoxylin, eosin and safran (HES) for routine microscopic examination. P16 and MIB-1 immunostainings were used to confirm the diagnosis and Human Papilloma Virus genotype determinations were done in all cases. All biopsies were reviewed by two pathologists, who were blinded to the original diagnosis and follow-up; in case of discrepancies the cases were re-reviewed with a double-head microscope by two pathologists (EG, JB) who also used the immunohistochemical stains and a consensus diagnosis was always obtained. The cases were diagnosed as follows: normal (n = 10) and CIN2-3 (n = 45).
2.4 Immunohistochemistry
The protocol for immunohistochemistry, type/manufacturer and dilution of the antibodies p16 and Ki67 has been described before [7]. For the epithelial biomarkers the following antibodies and dilutions were used: CK-13 (clone KS-1A3, Novocastra, Newcastle upon Tyne, UK) 1/200; CK-14 (clone LL002, Novocastra, Newcastle upon Tyne, UK) 1/40; pRB (clone 13A10, Novocastra, Newcastle upon Tyne, UK) 1/25; p53 (clone DO-7, DAKO, Glostrup, Denmark, S3002) 1/200; PhosphoHistone=PPH3 (Polyclonal Cell signaling solutions, Upstate #06-570; Lake Placid, NY, USA) 1/1500. The section adjacent to the sections used for immunostainings was cut and stained with H&E to ensure the presence of the same CIN lesion in all test sections (“sandwich-technique”).
2.5 ProteinChip SELDI-TOF MS analysis
Protein concentrations were assessed using a spectrophotometer (Thermo Scientific NanoDrop Spectrophotometer) measuring the absorbance at 280 nm. An equal amount of total protein (10 μg) was used. Samples were subjected to SELDI-TOF MS profiling according to the manufacturer’s instructions (Ciphergen Biosystems, Fremont, CA, USA). The biopsy supernatants were diluted 1:5 with 50 mM sodium acetate (pH 4.3) and then bound to a CM10 ProteinChip array. They were incubated for two hours at room temperature on a platform shaker, then washed twice with 50 mM sodium acetate buffer, followed by two times 1 μl of energy absorbing molecule (=EAM) solution (consisting of 50% saturated synaptic acid dissolved in 50% acetonitrile and 0.5% trifluoroacetic acid). Two replicates were prepared on different CM10 ProteinChips by two different analysts on two different days. The time-of-flight spectra were generated on the Protein Biological System II mass spectrometer reader (Ciphergen Biosystems, Fremont, CA, USA), using a laser intensity of 170, and a detector sensitivity of seven. Readings were optimized for low molecular weight (2–20 kDa). External mass calibration was performed daily.
2.6 Data analysis
The SELDI-TOF MS data analysis was performed in three steps: 1) peak detection, 2) selection of peaks with the highest discriminatory power and 3) building a multivariate model based on the selection in step 2. The peak detection was done using the Ciphergen Seldi software version 3.2 after internal and external mass calibration followed by normalization (TIC intensity) of all spectra as one group. The mass range from 2000 to 20000 Da contained the majority of the peptides/proteins in the samples, and was selected. Masses less than 2000 Da was excluded as these are known to contain adducts and artifacts from the EAM-solution and other chemical contaminants. The peak detection includes baseline subtraction, calibration of mass accuracy and automatic peak detection. Each spectrum was then assigned to one of two groups, normal or CIN2-3. To select peaks with the highest discriminatory power, the Biomarker Wizard (Ciphergen) was used for peak detection and clustering of all the spectra. This was done using a signal-to-noise (s/n) ratio of 5 and 15% of all spectra for the first pass detection and clustering, and an s/n ratio of 2 for the second pass. The cluster results were then imported into SPSS version 15 (SPSS Norway AS, Oslo, Norway), CART (Salford, San Diego, CA, USA) and MedCalc (MedCalc Software, Mariakerke, Belgium) for binary logistic regression analysis and Classification and Regression Tree analysis. A t-test was done, a correlation test using all 40 peaks followed by the development of a binary logistic regression model using the peaks that were found not to be correlated.
3 Results
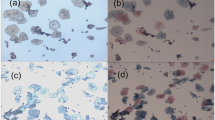
The median age of the patients at the time that the biopsies were obtained was 30.8 years (range 26–40) for the CIN2-3 group and for the normal biopsies group, 33.1 years (range 32–34) (these differences were not significant). The mean protein concentrations of the normal (1.00 mg/ml) and the CIN2-3 (1.09 mg/ml) samples were also not significantly different. We did evaluate the effect of different incubation times on the histology and immunohistochemistry on cervical biopsies. Figure 1 gives typical examples, showing that after 24 h incubation, there are no visible differences compared with 0 h incubation.
The peak detection and clustering process using Biomarker Wizard resulted in a total of 40 protein peaks. The development of a binary logistic model resulted in three peaks found to have independent value in discriminating the two groups. Figures 2 and 3 show zoom views of the differential peaks and examples of full spectra (m/z 2000–10000) for the 2 groups analyzed. As Fig. 2 shows, the peak with m/z 3430 is down-regulated in CIN2-3 samples compared to normal samples, while the peaks with m/z 5077 and 7794 up-regulated. The reproducibility between the technical replicates and between samples can be seen in Fig. 3 where the replicates for three different CIN2-3 samples are visualized as gel traces (gel view).
The reproducibility of the analysis was tested by comparing all 40 peaks between replicate 1 and 2. Only three of the 40 peaks were significant different between replicate 1 and replicate 2, i.e. m/z 3318 (p = 0.01), m/z 4023(p = 0.0001) and m/z 10885 (p = 0.03). All other peaks had p values higher than 0.10 and the three discriminatory peaks were not different at all (P > 0.80). When the multivariate analyses were repeated leaving out the peaks that were significantly different between the two replicates, the discriminatory results between the normal and CIN2-3 samples were the same as before.
Table 1 summarizes the 3 discriminant peaks. The average intensities, confidence intervals and the probabilities of no difference (t-test) between the normal and CIN2-3 groups assuming independency are shown. Figures 4, 5 and 6 shows the discrimination of the two groups using the three SELDI-TOF MS peaks as variables. By using the developed three-step binary logistic model, an overall correct classification of 88% was obtained. 14 of the 20 normal samples and 84 of 90 of the CIN2-3 samples were correctly classified.
4 Discussion
This study presents the results obtained from analyzing protein extracts obtained by a novel method by SELDI-TOF MS. We have shown that water-soluble proteins sampled from punch biopsies are promising to assist the diagnosis of normal and CIN2-3.
We found that 24 h immersion of cervical punch biopsies in RPMI at 4°C can be used to harvest 1 mg/ml water soluble proteins without compromising the routine immunohistochemical analysis of the tissue. Moreover, the water soluble proteins from the biopsies give good discrimination between samples with normal and CIN2-3 epithelia.
SELDI-TOF MS technology has been applied for several years to address the need for new markers for early cancer detection [19, 22–24] or for different lymphomas [25]. Most studies have compared diseased and healthy persons and have been useful for a better understanding of the development of several types of cancer. However, it is more difficult to develop biomarkers which can discriminate between the different development stages of a neoplasia as the differences are much smaller than between healthy and diseased persons. Yet, the model presented here shows that this still is possible with the technology used. CIN represents the first step in the development of cervical cancer through the three different CIN phases. In a future study, it is important to analyze whether SELDI-TOF MS or other mass spectrometric techniques can discriminate between CINs that will regress spontaneously from CINs that will persist or progress to invasive cancer. Furthermore, the protein collection method that was developed is of great interest for studying not only cervical neoplasia but it could also be applied to other organ tracts. The method of placing a biopsy in a cell culture medium for 24 h at 4°C is considerably longer than 1 h in PBS at 37°C used by Celis et al. [21], and enables the collection of proteins from a biopsy without interfering with the essential diagnostic information. The three peaks found to separate normal tissue and CIN2-3 lesions did not have the same m/z-values as those reported found from SELDI-TOF MS analysis of cell lysates of normal and invasive cervical cancer [26] or from plasma samples of CIN patients [27]. The protein concentrations in the samples were comparable to those found in samples from mucosal collection using sponges [28]. In addition, the RPMI immersion method has additional value to conventional FFPE analysis as it enables the collection and analysis of water soluble proteins which are not available from formalin-fixed paraffin-embedded tissue protein extracts. Proteins like cytokines and chemokines are collected in a volume enabling the analysis by different techniques like SELDI-TOF MS, LC-MS/MS or by ultrasensitive Elisa techniques like electrochemiluminescence.
The reviews published by Dijkstra et al. [29] and DeBock et al. [30] discuss several sources of variation that needs to be addressed. An essential aspect of a new laboratory test with possible diagnostic and therapeutic implications is the variation sources in all steps of the analysis [31]. These include: 1. Pre-analytical (a. each time getting the same samples, by using the same biopsy device; b. taking two samples from the same patient and put them in two different RPMI tubes; c. standardize the incubation conditions and time); 2. Analytical part (a. binding and cleanup on the SELDI-chip; b. reproducibility of the SELDI-TOF measurement of the same sample; c. take two samples from the same RPMI sample and test the reproducibility); 3. Post-analytical (a. peak processing and clustering; b. data analysis).
As to sampling device, we always used the same biopsy forceps. The samples were all taken by the same experienced gynaecologist. We also analyzed two nearby samples from 5 consecutive patients and compared the results. Two neighbouring biopsy samples in the cervix from the same patient also gave comparable results. We standardized the incubation conditions (RPMI, 4oC) and time (24 h).
As to the analytical part, all the technical aspects of the Seldi chip were standardized as described. We also tested the reproducibility of the same samples (see Fig. 3). Moreover, two different technicians analyzed the same samples, with the same results.
As to peak processing, clustering and data analysis, we used strictly protocolized standard operation procedures.
The results are promising but should be confirmed in a larger set of independent samples. In an ongoing study not published yet, a subset of supernatants has been characterized using LC-MS/MS. Further work to identify the candidate biomarkers found using this model will be done using LC-MS/MS (one- or multidimensional [32, 33]) of tryptic digests after fractionation by chromatography or two-dimensional gel electrophoresis [34]. An identification of the possible biomarker candidates found using this model will possibly give new insight to the mechanisms related to the Human Papilloma Virus infection.
5 Conclusions
Immersion of cervical punch biopsies for 24 h in RPMI tissue culture medium allows the analysis of water soluble proteins that can assist in the diagnosis of cervical intraepithelial neoplasia without interfering with the diagnosis. We conclude that the water soluble proteins from the biopsies provide good discrimination between normal and CIN2-3 samples. This is a promising strategy to study the dynamic behavior of CINs and hopefully will allow identification of regressive and non-regressive CIN2-3 lesions and a better indication of patient-tailored treatment.
References
A.E. Pollack, M.S. Balkin, L. Denny, Cervical cancer: a call for political will. Int. J. Gynaecol. Obstet. 94(3), 333–342 (2006)
J.M. Walboomers, M.V. Jacobs, M.M. Manos et al., Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189(1), 12–19 (1999)
R.P. Insinga, E.J. Dasbach, E.H. Elbasha, Epidemiologic natural history and clinical management of Human Papillomavirus (HPV) Disease: a critical and systematic review of the literature in the development of an HPV dynamic transmission model. BMC Infect. Dis. 9, 119 (2009)
J.P.A. Baak, A.J. Kruse, S.J. Robboy et al., Dynamic behavioural interpretation of cervical intraepithelial neoplasia with molecular biomarkers. J. Clin. Pathol. 59(10), 1017–1028 (2006)
P. Martin-Hirsch, E. Paraskevaidis, H. Kitchener et al., Surgery for cervical intraepithelial neoplasia. Cochrane Database Syst. Rev. 2, CD001318 (2000)
A.G. Ostor, Natural-history of cervical intraepithelial neoplasia - a critical review. Intern J Gynl Pathol 12(2), 186–192 (1993)
J.P.A. Baak, A.J. Kruse, Use of biomarkers in the evaluation of CIN grade and progression of early CIN. Meth. Mol. Med. 119, 85–99 (2005)
M.F. Addis, A. Tanca, D. Pagnozzi et al., Generation of high-quality protein extracts from formalin-fixed, paraffin-embedded tissues. Proteomics 9(15), 3815–3823 (2009)
M.F. Addis, A. Tanca, D. Pagnozzi et al., 2-D PAGE and MS analysis of proteins from formalin-fixed, paraffin-embedded tissues. Proteomics 9(18), 4329–4339 (2009)
B.L. Hood, M.M. Darfler, T.G. Guiel et al., Proteomic analysis of formalin-fixed prostate cancer tissue. Mol. Cell. Proteomics 4(11), 1741–1753 (2005)
B.L. Hood, T.P. Conrads, T.D. Veenstra, Unravelling the proteome of formalin-fixed paraffin-embedded tissue. Brief Funct Genomic Proteomic 5(2), 169–175 (2006)
P.E. Castle, T.M. Phillips, A. Hildesheim et al., Immune profiling of plasma and cervical secretions using recycling immunoaffinity chromatography. Cancer Epidemiol. Biomark. Prev. 12(12), 1449–1456 (2003)
S.H. Song, J.K. Lee, O.S. Seok et al., The relationship between cytokines and HPV-16, HPV-16 E6, E7, and high-risk HPV viral load in the uterine cervix. Gynecol. Oncol. 104(3), 732–738 (2007)
M.Y. Tjiong, N. van der Vange, J.S. ter Schegget et al., Cytokines in cervicovaginal washing fluid from patients with cervical neoplasia. Cytokine 14(6), 357–360 (2001)
P.E. Castle, A. Hildesheim, F.P. Bowman et al., Cervical concentrations of interleukin-10 and interleukin-12 do not correlate with plasma levels. J. Clin. Immunol. 22(1), 23–27 (2002)
M.Y. Tjiong, N. van der Vange, F.J.W. ten Kate, et al. Increased IL-6 and IL-8 levels in cervicovaginal secretions of patients with cervical cancer. 285–291 (1999)
J.A. Lieberman, A.B. Moscicki, J.L. Sumerel et al., Determination of cytokine protein levels in cervical mucus samples from young women by a multiplex immunoassay method and assessment of correlates. Clin Vacc Immun 15(1), 49–54 (2008)
K.K. Azar, M. Tani, H. Yasuda et al., Increased secretion patterns of interleukin-10 and tumor necrosis factor-alpha in cervical squamous intraepithelial lesions. Hum. Pathol. 35(11), 1376–1384 (2004)
E.F. Petricoin, L.A. Liotta, SELDI-TOF-based serum proteomic pattern diagnostics for early detection of cancer. Curr. Opin. Biotechnol. 15(1), 24–30 (2004)
E.F. Petricoin, D.K. Ornstein, L.A. Liotta, Clinical proteomics: Applications for prostate cancer biomarker discovery and detection. Urol. Oncol. 22(4), 322–328 (2004)
J.E. Celis, P. Gromov, T. Cabezòn et al., Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment. Mol. Cell. Proteomics 3, 327–344 (2004)
H.J. Issaq, T.P. Conrads, D.A. Prieto et al., SELDI-TOF MS for diagnostic proteomics. Anal. Chem. 75(7), 148A–155A (2003)
C.P. Paweletz, L.A. Liotta, E.F. Petricoin 3rd, New technologies for biomarker analysis of prostate cancer progression: Laser capture microdissection and tissue proteomics. Urology 57(4 Suppl 1), 160–163 (2001)
K.D. Rodland, Proteomics and cancer diagnosis: the potential of mass spectrometry. Clin. Biochem. 37(7), 579–583 (2004)
C. Jansen, K.M. Hebeda, M. Linkels et al., Protein profiling of B-cell lymphomas using tissue biopsies: a potential tool for small samples in pathology. Cell. Oncol. 30, 27–38 (2008)
Y.F. Wong, T.H. Cheung, K.W. Lo et al., Protein profiling of cervical cancer by protein-biochips: proteomic scoring to discriminate cervical cancer from normal cervix. Cancer Lett. 211(2), 227–234 (2004)
C.J. Piyathilake, D.K. Oelschlager, S. Meleth et al., Plasma protein profiles differ between women diagnosed with cervical intraepithelial neoplasia (CIN) 1 and 3. Cancer Inf. 2, 345–349 (2007)
G. Panicker, D.R. Lee, E.R. Unger, Optimization of SELDI-TOF protein profiling for analysis of cervical mucous. J. Proteomics 71(6), 637–646 (2009)
M. Dijkstra, R.J. Vonk, R.C. Jansen, SELDI-TOF mass spectra: a review on sources of variation. J. Chromatogr. B 847, 12–23 (2007)
M. De Bock, D. de Seny, M.-A. Meuwis et al., Challenges for biomarker discovery in body fluids using SELDI-TOF MS. J. Biomed. Biotechnol. 2010, 906082 (2009)
J.P.A. Baak, The framework of pathology: good laboratory practice by quantitative and molecular methods. J. Pathol. 198, 277–283 (2002)
H.J. Issaq, K.C. Chan, G.M. Janini et al., Multidimensional separation of peptides for effective proteomic analysis. J Chromatogr B Analyt Technol Biomed Life Sci 817(1), 35–47 (2005)
W.H. McDonald, R. Ohi, D.T. Miyamoto et al., Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 219(1), 245–251 (2002)
A. Gorg, W. Weiss, M.J. Dunn, Current two-dimensional electrophoresis technology for proteomics. Proteomics 4(12), 3665–3685 (2004)
Acknowledgements
Funding for this research was provided by Folke Hermansen Foundation.
Disclusure/conflict of interest
The authors declare that they have no competing interests.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Kai-Erik Uleberg and Ane Cecilie Munk contributed equally to this work
This study was financially supported by a grant from the Folke Hermansen Foundation in 2007
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Uleberg, KE., Munk, A.C., Skaland, I. et al. A protein profile study to discriminate CIN lesions from normal cervical epithelium. Cell Oncol. 34, 443–450 (2011). https://doi.org/10.1007/s13402-011-0047-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-011-0047-3